Identification and Validation of New DNA-PKcs Inhibitors through High-Throughput Virtual Screening and Experimental Verification
- PMID: 39063224
- PMCID: PMC11277333
- DOI: 10.3390/ijms25147982
Identification and Validation of New DNA-PKcs Inhibitors through High-Throughput Virtual Screening and Experimental Verification
Abstract
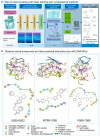
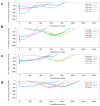
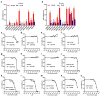
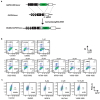
DNA-PKcs is a crucial protein target involved in DNA repair and response pathways, with its abnormal activity closely associated with the occurrence and progression of various cancers. In this study, we employed a deep learning-based screening and molecular dynamics (MD) simulation-based pipeline, identifying eight candidates for DNA-PKcs targets. Subsequent experiments revealed the effective inhibition of DNA-PKcs-mediated cell proliferation by three small molecules (5025-0002, M769-1095, and V008-1080). These molecules exhibited anticancer activity with IC50 (inhibitory concentration at 50%) values of 152.6 μM, 30.71 μM, and 74.84 μM, respectively. Notably, V008-1080 enhanced homology-directed repair (HDR) mediated by CRISPR/Cas9 while inhibiting non-homologous end joining (NHEJ) efficiency. Further investigations into the structure-activity relationships unveiled the binding sites and critical interactions between these small molecules and DNA-PKcs. This is the first application of DeepBindGCN_RG in a real drug screening task, and the successful discovery of a novel DNA-PKcs inhibitor demonstrates its efficiency as a core component in the screening pipeline. Moreover, this study provides important insights for exploring novel anticancer therapeutics and advancing the development of gene editing techniques by targeting DNA-PKcs.
Keywords: CRISPR/Cas9; DNA-PKcs; HDR; anticancer activity; deep learning; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
In-silico screening for DNA-dependent protein kinase (DNA-PK) inhibitors: Combined homology modeling, docking, molecular dynamic study followed by biological investigation.Biomed Pharmacother. 2016 Oct;83:693-703. doi: 10.1016/j.biopha.2016.07.044. Epub 2016 Jul 28. Biomed Pharmacother. 2016. PMID: 27470570
-
A novel small molecule inhibitor of the DNA repair protein Ku70/80.DNA Repair (Amst). 2016 Jul;43:98-106. doi: 10.1016/j.dnarep.2016.03.014. Epub 2016 Apr 7. DNA Repair (Amst). 2016. PMID: 27130816
-
DNA-PKcs inhibition sensitizes cancer cells to carbon-ion irradiation via telomere capping disruption.PLoS One. 2013 Aug 27;8(8):e72641. doi: 10.1371/journal.pone.0072641. eCollection 2013. PLoS One. 2013. PMID: 24013362 Free PMC article.
-
DNA-PKcs: A promising therapeutic target in human hepatocellular carcinoma?DNA Repair (Amst). 2016 Nov;47:12-20. doi: 10.1016/j.dnarep.2016.10.004. Epub 2016 Oct 15. DNA Repair (Amst). 2016. PMID: 27789167 Review.
-
Advance trends in targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems.Bioimpacts. 2022;12(4):371-391. doi: 10.34172/bi.2022.23871. Epub 2022 Jun 22. Bioimpacts. 2022. PMID: 35975201 Free PMC article. Review.
Cited by
-
Bevacizumab in ovarian cancer therapy: current advances, clinical challenges, and emerging strategies.Front Bioeng Biotechnol. 2025 May 15;13:1589841. doi: 10.3389/fbioe.2025.1589841. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40474872 Free PMC article. Review.
-
ITGAX promotes gastric cancer progression via epithelial-mesenchymal transition pathway.Front Pharmacol. 2025 Jan 8;15:1536478. doi: 10.3389/fphar.2024.1536478. eCollection 2024. Front Pharmacol. 2025. PMID: 39845786 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

