A New Casjensviridae Bacteriophage Isolated from Hospital Sewage for Inactivation of Biofilms of Carbapenem Resistant Klebsiella pneumoniae Clinical Isolates
- PMID: 39065601
- PMCID: PMC11280391
- DOI: 10.3390/pharmaceutics16070904
A New Casjensviridae Bacteriophage Isolated from Hospital Sewage for Inactivation of Biofilms of Carbapenem Resistant Klebsiella pneumoniae Clinical Isolates
Abstract
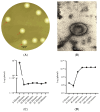
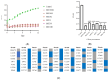
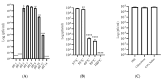
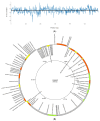
Klebsiella pneumoniae, a member of the ESKAPE pathogen group, is a prominent cause of hospital-acquired infections. The WHO has recognized carbapenem-resistant K. pneumoniae as a critical-one priority pathogen. These resilient superbugs have the ability to form biofilms and present a significant global threat. In the present study, we isolated and characterized a bacteriophage SAKp02, from hospital sewage, infectious to carbapenem-resistant K. pneumoniae patient isolates. SAKp02 could infect 43 of 72 clinical isolates, indicating a broad host spectrum. Whole genome analysis classified SAKp02 within the family Casjensviridae, with a 59,343 bp genome encoding 82 ORFs. Comparative genomic analysis revealed significant differences between SAKp02 and its closest viruses, indicating a distinct genetic makeup positioning it as a novel phage strain within the lineage. The SAKp02 genome comprises bacteriolytic enzymes, including holin, endolysin, and phage depolymerase, crucial for bacterial lysis and biofilm disruption. It reduced biofilm biomass by over threefold compared to the control and eradicated 99% of viable cells within a 4 h treatment period. Scanning electron microscopy corroborated the ability of the phage to dismantle biofilm matrices and lyse bacterial cells. Safe and effective treatments are warranted, and hence, the fully characterized lytic phages with therapeutic potential against drug-resistant clinical isolates of bacteria are needed. Our study is the first to report the antibacterial and antibiofilm activity of Casjensviridae phages, and our discovery of a novel K. pneumoniae phage broadens the arsenal against the bacteria.
Keywords: K. pneumoniae; alternative to antibiotics; antibiofilm strategy; antimicrobial resistance; biofilm bacteriophage.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Mohd Asri N.A., Ahmad S., Mohamud R., Mohd Hanafi N., Mohd Zaidi N.F., Irekeola A.A., Shueb R.H., Yee L.C., Mohd Noor N., Mustafa F.H., et al. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics. 2021;10:1508. doi: 10.3390/antibiotics10121508. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

