A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern
- PMID: 39065742
- PMCID: PMC11279616
- DOI: 10.3390/ph17070891
A Dynamic and Effective Peptide-Based Strategy for Promptly Addressing Emerging SARS-CoV-2 Variants of Concern
Abstract
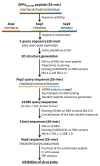
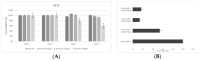
Genomic surveillance based on sequencing the entire genetic code of SARS-CoV-2 involves monitoring and studying genetic changes and variations in disease-causing organisms such as viruses and bacteria. By tracing the virus, it is possible to prevent epidemic spread in the community, ensuring a 'precision public health' strategy. A peptide-based design was applied to provide an efficacious strategy that is able to counteract any emerging viral variant of concern dynamically and promptly to affect the outcomes of a pandemic at an early stage while waiting for the production of the anti-variant-specific vaccine, which require longer times. The inhibition of the interaction between the receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and one of the cellular receptors (DPP4) that its receptors routinely bind to infect human cells is an intriguing therapeutic approach to prevent the virus from entering human cells. Among the other modalities developed for this purpose, peptides surely offer unique advantages, including ease of synthesis, serum stability, low immunogenicity and toxicity, and small production and distribution chain costs. Here, we obtained a potent new inhibitor based on the rearrangement of a previously identified peptide that has been rationally designed on a cell dipeptidyl peptidase 4 (DPP4) sequence, a ubiquitous membrane protein known to bind the RBD-SPIKE domain of the virus. This novel peptide (named DPP4-derived), conceived as an endogenous "drug", is capable of targeting the latest tested variants with a high affinity, reducing the VSV* DG-Fluc pseudovirus Omicron's infection capacity by up to 14%, as revealed by in vitro testing in human Calu-3 cells. Surface plasmon resonance (SPR) confirmed the binding affinity of the new DPP4-derived peptide with Omicron variant RBD.
Keywords: RBD variants; SARS-CoV-2; peptides; protein–protein interaction (PPI); pseudovirus; surface plasmon resonance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
Design of a bifunctional pan-sarbecovirus entry inhibitor targeting the cell receptor and viral fusion protein.J Virol. 2023 Aug 31;97(8):e0019223. doi: 10.1128/jvi.00192-23. Epub 2023 Aug 14. J Virol. 2023. PMID: 37578234 Free PMC article.
-
Novel Affibody Molecules Specifically Bind to SARS-CoV-2 Spike Protein and Efficiently Neutralize Delta and Omicron Variants.Microbiol Spectr. 2023 Feb 14;11(1):e0356222. doi: 10.1128/spectrum.03562-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511681 Free PMC article.
-
Surface charge changes in spike RBD mutations of SARS-CoV-2 and its variant strains alter the virus evasiveness via HSPGs: A review and mechanistic hypothesis.Front Public Health. 2022 Aug 24;10:952916. doi: 10.3389/fpubh.2022.952916. eCollection 2022. Front Public Health. 2022. PMID: 36091499 Free PMC article. Review.
-
Emergence, evolution, and vaccine production approaches of SARS-CoV-2 virus: Benefits of getting vaccinated and common questions.Saudi J Biol Sci. 2022 Apr;29(4):1981-1997. doi: 10.1016/j.sjbs.2021.12.020. Epub 2021 Dec 13. Saudi J Biol Sci. 2022. PMID: 34924802 Free PMC article. Review.
Cited by
-
SARS-CoV-2 Variants: Genetic Insights, Epidemiological Tracking, and Implications for Vaccine Strategies.Int J Mol Sci. 2025 Jan 31;26(3):1263. doi: 10.3390/ijms26031263. Int J Mol Sci. 2025. PMID: 39941026 Free PMC article. Review.
-
Rerouting therapeutic peptides and unlocking their potential against SARS-CoV2.3 Biotech. 2025 May;15(5):116. doi: 10.1007/s13205-025-04270-0. Epub 2025 Apr 4. 3 Biotech. 2025. PMID: 40191455 Review.
References
-
- World Health Organization (WHO) Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. [(accessed on 5 May 2023)]. Available online: www.who.int/news-room/fact-sheets/detail/coronavirus-disease-(covid-19)
-
- Atella V., Scandizzo P.L. Chapter 1—The origins of infections: Some basic concepts. In: Atella V., Scandizzo P.L., editors. The COVID-19 Disruption and the Global Health Challenge. Academic Press; Cambridge, MA, USA: 2024. pp. 3–20.
-
- Atella V., Scandizzo P.L. Chapter 11—How to manage the risk of new pandemics. In: Atella V., Scandizzo P.L., editors. The COVID-19 Disruption and the Global Health Challenge. Academic Press; Cambridge, MA, USA: 2024. pp. 409–438.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

