Chandipura Virus Forms Cytoplasmic Inclusion Bodies through Phase Separation and Proviral Association of Cellular Protein Kinase R and Stress Granule Protein TIA-1
- PMID: 39066190
- PMCID: PMC11281494
- DOI: 10.3390/v16071027
Chandipura Virus Forms Cytoplasmic Inclusion Bodies through Phase Separation and Proviral Association of Cellular Protein Kinase R and Stress Granule Protein TIA-1
Abstract
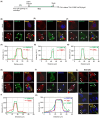
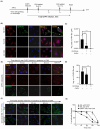
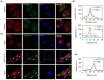
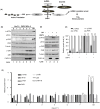
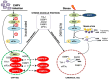
Negative-strand RNA viruses form cytoplasmic inclusion bodies (IBs) representing virus replication foci through phase separation or biomolecular condensation of viral and cellular proteins, as a hallmark of their infection. Alternatively, mammalian cells form stalled mRNA containing antiviral stress granules (SGs), as a consequence of phosphorylation of eukaryotic initiation factor 2α (eIF2α) through condensation of several RNA-binding proteins including TIA-1. Whether and how Chandipura virus (CHPV), an emerging human pathogen causing influenza-like illness, coma and death, forms IBs and evades antiviral SGs remain unknown. By confocal imaging on CHPV-infected Vero-E6 cells, we found that CHPV infection does not induce formation of distinct canonical SGs. Instead, CHPV proteins condense and co-localize together with SG proteins to form heterogeneous IBs, which ensued independent of the activation of eIF2α and eIF2α kinase, protein kinase R (PKR). Interestingly, siRNA-mediated depletion of PKR or TIA-1 significantly decreased viral transcription and virion production. Moreover, CHPV infection also caused condensation and recruitment of PKR to IBs. Compared to SGs, IBs exhibited significant rapidity in disassembly dynamics. Altogether, our study demonstrating that CHPV replication co-optimizes with SG proteins and revealing an unprecedented proviral role of TIA-1/PKR may have implications in understanding the mechanisms regulating CHPV-IB formation and designing antiviral therapeutics. Importance: CHPV is an emerging tropical pathogen reported to cause acute influenza-like illness and encephalitis in children with a very high mortality rate of ~70%. Lack of vaccines and an effective therapy against CHPV makes it a potent pathogen for causing an epidemic in tropical parts of globe. Given these forewarnings, it is of paramount importance that CHPV biology must be understood comprehensively. Targeting of host factors offers several advantages over targeting the viral components due to the generally higher mutation rate in the viral genome. In this study, we aimed at understanding the role of SGs forming cellular RNA-binding proteins in CHPV replication. Our study helps understand participation of cellular factors in CHPV replication and could help develop effective therapeutics against the virus.
Keywords: Chandipura virus; inclusion bodies; phase separation; protein kinase R; stress granules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rao B.L., Basu A., Wairagkar N.S., Gore M.M., Arankalle V.A., Thakare J.P., Jadi R.S., Rao K.A., Mishra A.C. A large outbreak of acute encephalitis with high fatality rate in children in Andhra Pradesh, India, in 2003, associated with Chandipura virus. Lancet. 2004;364:869–874. doi: 10.1016/S0140-6736(04)16982-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

