Protein C Pretreatment Protects Endothelial Cells from SARS-CoV-2-Induced Activation
- PMID: 39066212
- PMCID: PMC11281670
- DOI: 10.3390/v16071049
Protein C Pretreatment Protects Endothelial Cells from SARS-CoV-2-Induced Activation
Abstract
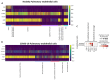
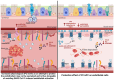
SARS-CoV-2 can induce vascular dysfunction and thrombotic events in patients with severe COVID-19; however, the cellular and molecular mechanisms behind these effects remain largely unknown. In this study, we used a combination of experimental and in silico approaches to investigate the role of PC in vascular and thrombotic events in COVID-19. Single-cell RNA-sequencing data from patients with COVID-19 and healthy subjects were obtained from the publicly available Gene Expression Omnibus (GEO) repository. In addition, HUVECs were treated with inactive protein C before exposure to SARS-CoV-2 infection or a severe COVID-19 serum. An RT-qPCR array containing 84 related genes was used, and the candidate genes obtained were evaluated. Activated protein C levels were measured using an ELISA kit. We identified at the single-cell level the expression of several pro-inflammatory and pro-coagulation genes in endothelial cells from the patients with COVID-19. Furthermore, we demonstrated that exposure to SARS-CoV-2 promoted transcriptional changes in HUVECs that were partly reversed by the activated protein C pretreatment. We also observed that the serum of severe COVID-19 had a significant amount of activated protein C that could protect endothelial cells from serum-induced activation. In conclusion, activated protein C protects endothelial cells from pro-inflammatory and pro-coagulant effects during exposure to the SARS-CoV-2 virus.
Keywords: SARS-CoV-2; bioinformatics; blood coagulation disorders; endothelial cell; inflammation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. - DOI - PMC - PubMed
-
- Charfeddine S., Ibn Hadj Amor H., Jdidi J., Torjmen S., Kraiem S., Hammami R., Bahloul A., Kallel N., Moussa N., Touil I. Long COVID-19 syndrome: Is it related to microcirculation and endothelial dysfunction? Insights from TUN-EndCOV study. Front. Cardiovasc. Med. 2021;8:745758. doi: 10.3389/fcvm.2021.745758. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

