Comparative Proteomic Analysis of Huh7 Cells Transfected with Sub-Saharan African Hepatitis B Virus (Sub)genotypes Reveals Potential Oncogenic Factors
- PMID: 39066215
- PMCID: PMC11281506
- DOI: 10.3390/v16071052
Comparative Proteomic Analysis of Huh7 Cells Transfected with Sub-Saharan African Hepatitis B Virus (Sub)genotypes Reveals Potential Oncogenic Factors
Abstract
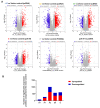
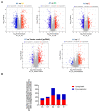
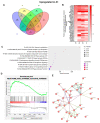
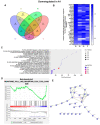
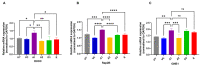
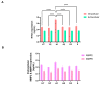
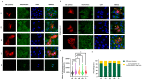
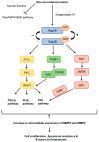
In sub-Saharan Africa (SSA), the (sub)genotypes A1, D3, and E of the hepatitis B virus (HBV) prevail. Individuals infected with subgenotype A1 have a 4.5-fold increased risk of HCC compared to those infected with other (sub)genotypes. The effect of (sub)genotypes on protein expression and host signalling has not been studied. Mass spectrometry was used to analyse the proteome of Huh7 cells transfected with replication-competent clones. Proteomic analysis revealed significantly differentially expressed proteins between SSA (sub)genotypes. Different (sub)genotypes have the propensity to dysregulate specific host signalling pathways. Subgenotype A1 resulted in dysregulation within the Ras pathway. Ras-associated protein, RhoC, was significantly upregulated in cells transfected with subgenotype A1 compared to those transfected with other (sub)genotypes, on both a proteomic (>1.5-fold) and mRNA level (p < 0.05). Two of the main cellular signalling pathways involving RHOC, MAPK and PI3K/Akt/mTOR, regulate cell growth, motility, and survival. Downstream signalling products of these pathways have been shown to increase MMP2 and MMP9 expression. An extracellular MMP2 and MMP9 ELISA revealed a non-significant increase in MMP2 and MMP9 in the cells transfected with A1 compared to the other (sub)genotypes (p < 0.05). The upregulated Ras-associated proteins have been implicated as oncoproteins in various cancers and could contribute to the increased hepatocarcinogenic potential of A1.
Keywords: (sub)genotypes; hepatitis B virus; hepatocellular carcinoma; oncogenic pathways; viral oncogenesis.
Conflict of interest statement
S.S. has a commercial interest in ReSyn Biosciences which supplied the magnetic microspheres used during mass spectrometry sample preparation. The authors have no additional conflicts of interest to declare.
Figures


References
-
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Bsc M.F.B., Me J.F., Soerjomataram M.I., et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. - DOI - PubMed
-
- WHO . Global Hepatitis Report 2024: Action for Access in Low- and Middle-Income Countries. World Health Organization; Geneva, Switzerland: 2024.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

