Global Update on Measles Molecular Epidemiology
- PMID: 39066448
- PMCID: PMC11281501
- DOI: 10.3390/vaccines12070810
Global Update on Measles Molecular Epidemiology
Abstract
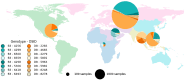
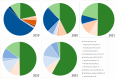
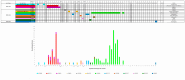
Molecular surveillance of circulating measles variants serves as a line of evidence for the absence of endemic circulation and provides a means to track chains of transmission. Molecular surveillance for measles (genotyping) is based on the sequence of 450 nucleotides at the end of the nucleoprotein coding region (N450) of the measles genome. Genotyping was established in 1998 and, with over 50,000 sequence submissions to the Measles Nucleotide Surveillance database, has proven to be an effective resource for countries attempting to trace pathways of transmission. This review summarizes the tools used for the molecular surveillance of measles and describes the challenge posed by the decreased number of circulating measles genotypes. The Global Measles and Rubella Laboratory Network addressed this challenge through the development of new tools such as named strains and distinct sequence identifiers that analyze the diversity within the currently circulating genotypes. The advantages and limitations of these approaches are discussed, together with the need to generate additional sequence data including whole genome sequences to ensure the continued utility of strain surveillance for measles.
Keywords: elimination; genotyping; measles; molecular surveillance.
Conflict of interest statement
Author Derek Hart was employed by the company ASRT, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Measles Overview. [(accessed on 9 May 2024)]. Available online: https://www.who.int/health-topics/measles.
-
- World Health Organization Monitoring progress towards measles elimination. Wkly. Epidemiol. Rec. 2010;85:490–494. - PubMed
-
- World Health Organization Guidance for evaluating progress towards elimination of measles and rubella. Wkly. Epidemiol. Rec. 2018;93:544–552.
-
- World Health Organization Standardization of the nomenclature for describing the genetic characteristics of wild-type measles viruses. Wkly. Epidemiol. Rec. 1998;73:265–271. - PubMed
-
- World Health Organization Update of the nomenclature for describing the genetic characteristics of wild-type measles viruses: New genotypes and reference strains. Wkly. Epidemiol. Rec. 2003;78:229–232. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

