Folate-mediated transgenerational inheritance of sperm DNA methylation patterns correlate with spinal axon regeneration
- PMID: 39066680
- PMCID: PMC11285217
- DOI: 10.1080/15592294.2024.2380930
Folate-mediated transgenerational inheritance of sperm DNA methylation patterns correlate with spinal axon regeneration
Erratum in
-
Correction.Epigenetics. 2024 Dec;19(1):2388387. doi: 10.1080/15592294.2024.2388387. Epub 2024 Aug 4. Epigenetics. 2024. PMID: 39097935 Free PMC article. No abstract available.
Abstract
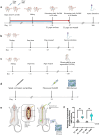
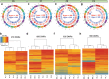
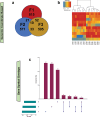
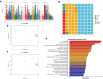
In mammals, the molecular mechanisms underlying transgenerational inheritance of phenotypic traits in serial generations of progeny after ancestral environmental exposures, without variation in DNA sequence, remain elusive. We've recently described transmission of a beneficial trait in rats and mice, in which F0 supplementation of methyl donors, including folic acid, generates enhanced axon regeneration after sharp spinal cord injury in untreated F1 to F3 progeny linked to differential DNA methylation levels in spinal cord tissue. To test whether the transgenerational effect of folic acid is transmitted via the germline, we performed whole-genome methylation sequencing on sperm DNA from F0 mice treated with either folic acid or vehicle control, and their F1, F2, and F3 untreated progeny. Transgenerational differentially methylated regions (DMRs) are observed in each consecutive generation and distinguish folic acid from untreated lineages, predominate outside of CpG islands and in regions of the genome that regulate gene expression, including promoters, and overlap at both the differentially methylated position (DMP) and gene levels. These findings indicate that molecular changes between generations are caused by ancestral folate supplementation. In addition, 29,719 DMPs exhibit serial increases or decreases in DNA methylation levels in successive generations of untreated offspring, correlating with a serial increase in the phenotype across generations, consistent with a 'wash-in' effect. Sibship-specific DMPs annotate to genes that participate in axon- and synapse-related pathways.
Keywords: CNS regeneration; Folic acid; spinal cord injury; transgenerational epigenetic inheritance; whole genome methylation sequencing (WGMS).
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
