Isogenic patient-derived organoids reveal early neurodevelopmental defects in spinal muscular atrophy initiation
- PMID: 39067446
- PMCID: PMC11384962
- DOI: 10.1016/j.xcrm.2024.101659
Isogenic patient-derived organoids reveal early neurodevelopmental defects in spinal muscular atrophy initiation
Abstract
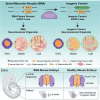
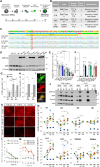
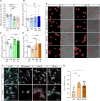
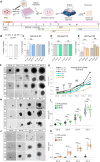
Whether neurodevelopmental defects underlie postnatal neuronal death in neurodegeneration is an intriguing hypothesis only recently explored. Here, we focus on spinal muscular atrophy (SMA), a neuromuscular disorder caused by reduced survival of motor neuron (SMN) protein levels leading to spinal motor neuron (MN) loss and muscle wasting. Using the first isogenic patient-derived induced pluripotent stem cell (iPSC) model and a spinal cord organoid (SCO) system, we show that SMA SCOs exhibit abnormal morphological development, reduced expression of early neural progenitor markers, and accelerated expression of MN progenitor and MN markers. Longitudinal single-cell RNA sequencing reveals marked defects in neural stem cell specification and fewer MNs, favoring mesodermal progenitors and muscle cells, a bias also seen in early SMA mouse embryos. Surprisingly, SMN2-to-SMN1 conversion does not fully reverse these developmental abnormalities. These suggest that early neurodevelopmental defects may underlie later MN degeneration, indicating that postnatal SMN-increasing interventions might not completely amend SMA pathology in all patients.
Keywords: isogenic SMA model; neurodevelopmental defects; neuromesodermal progenitors; organoids; spinal cord.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

