Automated preparation of plasma lipids, metabolites, and proteins for LC/MS-based analysis of a high-fat diet in mice
- PMID: 39067520
- PMCID: PMC11399584
- DOI: 10.1016/j.jlr.2024.100607
Automated preparation of plasma lipids, metabolites, and proteins for LC/MS-based analysis of a high-fat diet in mice
Abstract
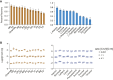
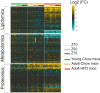
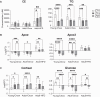
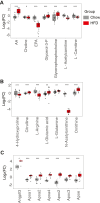
Blood plasma is one of the most commonly analyzed and easily accessible biological samples. Here, we describe an automated liquid-liquid extraction platform that generates accurate, precise, and reproducible samples for metabolomic, lipidomic, and proteomic analyses from a single aliquot of plasma while minimizing hands-on time and avoiding contamination from plasticware. We applied mass spectrometry to examine the metabolome, lipidome, and proteome of 90 plasma samples to determine the effects of age, time of day, and a high-fat diet in mice. From 25 μl of mouse plasma, we identified 907 lipid species from 16 different lipid classes and subclasses, 233 polar metabolites, and 344 proteins. We found that the high-fat diet induced only mild changes in the polar metabolome, upregulated apolipoproteins, and induced substantial shifts in the lipidome, including a significant increase in arachidonic acid and a decrease in eicosapentaenoic acid content across all lipid classes.
Keywords: MTBE-LLE; apolipoprotein; cholesterol/trafficking; dyslipidemias; lipidomics; multiomics; omega-3 fatty acid; phospholipids/metabolism.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

