A bacteriocin expression platform for targeting pathogenic bacterial species
- PMID: 39068147
- PMCID: PMC11283563
- DOI: 10.1038/s41467-024-50591-8
A bacteriocin expression platform for targeting pathogenic bacterial species
Abstract
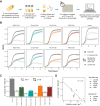
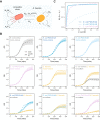
Bacteriocins are antimicrobial peptides that are naturally produced by many bacteria. They hold great potential in the fight against antibiotic resistant bacteria, including ESKAPE pathogens. Engineered live biotherapeutic products (eLBPs) that secrete bacteriocins can be created to deliver targeted bacteriocin production. Here we develop a modular bacteriocin secretion platform that can be used to express and secrete multiple bacteriocins from non-pathogenic Escherichia coli host strains. As a proof of concept we create Enterocin A (EntA) and Enterocin B (EntB) secreting strains that show strong antimicrobial activity against Enterococcus faecalis and Enterococcus faecium in vitro, and characterise this activity in both solid culture and liquid co-culture. We then develop a Lotka-Volterra model that can be used to capture the interactions of these competitor strains. We show that simultaneous exposure to EntA and EntB can delay Enterococcus growth. Our system has the potential to be used as an eLBP to secrete additional bacteriocins for the targeted killing of pathogenic bacteria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Bai, X. et al. Engineering the gut microbiome. Nat. Rev. Bioeng.1, 665–679 (2023).10.1038/s44222-023-00072-2 - DOI
MeSH terms
Substances
Grants and funding
- EP/W004674/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- EP/X026892/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- EP/W004674/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- EP/W004674/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- MR/W025655/1/RCUK | Medical Research Council (MRC)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

