Increased AID results in mutations at the CRLF2 locus implicated in Latin American ALL health disparities
- PMID: 39068148
- PMCID: PMC11283463
- DOI: 10.1038/s41467-024-50537-0
Increased AID results in mutations at the CRLF2 locus implicated in Latin American ALL health disparities
Abstract
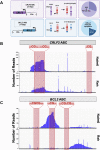
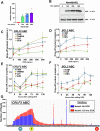
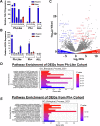
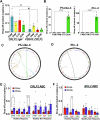
Activation-induced cytidine deaminase (AID) is a B cell-specific mutator required for antibody diversification. However, it is also implicated in the etiology of several B cell malignancies. Evaluating the AID-induced mutation load in patients at-risk for certain blood cancers is critical in assessing disease severity and treatment options. We have developed a digital PCR (dPCR) assay that allows us to quantify mutations resulting from AID modification or DNA double-strand break (DSB) formation and repair at sites known to be prone to DSBs. Implementation of this assay shows that increased AID levels in immature B cells increase genome instability at loci linked to chromosomal translocation formation. This includes the CRLF2 locus that is often involved in translocations associated with a subtype of acute lymphoblastic leukemia (ALL) that disproportionately affects Hispanics, particularly those with Latin American ancestry. Using dPCR, we characterize the CRLF2 locus in B cell-derived genomic DNA from both Hispanic ALL patients and healthy Hispanic donors and found increased mutations in both, suggesting that vulnerability to DNA damage at CRLF2 may be driving this health disparity. Our ability to detect and quantify these mutations will potentiate future risk identification, early detection of cancers, and reduction of associated cancer health disparities.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Update of
-
Increased AID Results in Mutations at the CRLF2 Locus Implicated in Latin American ALL Health Disparities.Res Sq [Preprint]. 2023 Sep 11:rs.3.rs-3332673. doi: 10.21203/rs.3.rs-3332673/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Jul 27;15(1):6331. doi: 10.1038/s41467-024-50537-0. PMID: 37790327 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 CA244519/CA/NCI NIH HHS/United States
- R01CA259370/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R37CA266042/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01 CA276470/CA/NCI NIH HHS/United States
- DP1 DK130640/DK/NIDDK NIH HHS/United States
- T32 CA009054/CA/NCI NIH HHS/United States
- R37 CA266042/CA/NCI NIH HHS/United States
- IRG-16-187-13/American Cancer Society (American Cancer Society, Inc.)
- P20 CA253251/CA/NCI NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- R21AR082841/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01CA244519/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01CA276470/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- DP1DK130640/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P20 CA253254/CA/NCI NIH HHS/United States
- R01 CA259370/CA/NCI NIH HHS/United States
- R21 AR082842/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

