Adenosine triggers early astrocyte reactivity that provokes microglial responses and drives the pathogenesis of sepsis-associated encephalopathy in mice
- PMID: 39068155
- PMCID: PMC11283516
- DOI: 10.1038/s41467-024-50466-y
Adenosine triggers early astrocyte reactivity that provokes microglial responses and drives the pathogenesis of sepsis-associated encephalopathy in mice
Erratum in
-
Author Correction: Adenosine triggers early astrocyte reactivity that provokes microglial responses and drives the pathogenesis of sepsis-associated encephalopathy in mice.Nat Commun. 2024 Sep 18;15(1):8200. doi: 10.1038/s41467-024-52497-x. Nat Commun. 2024. PMID: 39294127 Free PMC article. No abstract available.
Abstract
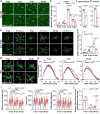
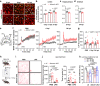
Molecular pathways mediating systemic inflammation entering the brain parenchyma to induce sepsis-associated encephalopathy (SAE) remain elusive. Here, we report that in mice during the first 6 hours of peripheral lipopolysaccharide (LPS)-evoked systemic inflammation (6 hpi), the plasma level of adenosine quickly increased and enhanced the tone of central extracellular adenosine which then provoked neuroinflammation by triggering early astrocyte reactivity. Specific ablation of astrocytic Gi protein-coupled A1 adenosine receptors (A1ARs) prevented this early reactivity and reduced the levels of inflammatory factors (e.g., CCL2, CCL5, and CXCL1) in astrocytes, thereby alleviating microglial reaction, ameliorating blood-brain barrier disruption, peripheral immune cell infiltration, neuronal dysfunction, and depression-like behaviour in the mice. Chemogenetic stimulation of Gi signaling in A1AR-deficent astrocytes at 2 and 4 hpi of LPS injection could restore neuroinflammation and depression-like behaviour, highlighting astrocytes rather than microglia as early drivers of neuroinflammation. Our results identify early astrocyte reactivity towards peripheral and central levels of adenosine as an important pathway driving SAE and highlight the potential of targeting A1ARs for therapeutic intervention.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures






References
-
- van der Poll, T., van de Veerdonk, F. L., Scicluna, B. P. & Netea, M. G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol.17, 407–420 (2017). - PubMed
-
- Duan, L. et al. PDGFRβ cells rapidly relay inflammatory signal from the circulatory system to neurons via chemokine CCL2. Neuron100, 183–200.e188 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- 462650276/Deutsche Forschungsgemeinschaft (German Research Foundation)
- KI 503/14-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1158/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SPP 1757/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 10.21.1.021MN/Fritz Thyssen Stiftung (Fritz Thyssen Foundation)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

