Combined Inhibition of PI3K and STAT3 signaling effectively inhibits bladder cancer growth
- PMID: 39068158
- PMCID: PMC11283499
- DOI: 10.1038/s41389-024-00529-y
Combined Inhibition of PI3K and STAT3 signaling effectively inhibits bladder cancer growth
Abstract
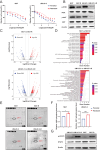
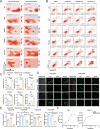
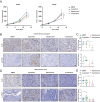
Bladder cancer is characterized by aberrant activation of the phosphatidylinositol-3-OH kinase (PI3K) signaling, underscoring the significance of directing therapeutic efforts toward the PI3K pathway as a promising strategy. In this study, we discovered that PI3K serves as a potent therapeutic target for bladder cancer through a high-throughput screening of inhibitory molecules. The PI3K inhibitor demonstrated a robust anti-tumor efficacy, validated both in vitro and in vivo settings. Nevertheless, the feedback activation of JAK1-STAT3 signaling reinstated cell and organoid survival, leading to resistance against the PI3K inhibitor. Mechanistically, the PI3K inhibitor suppresses PTPN11 expression, a negative regulator of the JAK-STAT pathway, thereby activating STAT3. Conversely, restoration of PTPN11 enhances the sensitivity of cancer cells to the PI3K inhibitor. Simultaneous inhibition of both PI3K and STAT3 with small-molecule inhibitors resulted in sustained tumor regression in patient-derived bladder cancer xenografts. These findings advocate for a combinational therapeutic approach targeting both PI3K and STAT3 pathways to achieve enduring cancer eradication in vitro and in vivo, underscoring their promising therapeutic efficacy for treating bladder cancer.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
Grants and funding
- U22A20331/National Natural Science Foundation of China (National Science Foundation of China)
- 82273414/National Natural Science Foundation of China (National Science Foundation of China)
- 82003000/National Natural Science Foundation of China (National Science Foundation of China)
- 82102736/National Natural Science Foundation of China (National Science Foundation of China)
- RCBS20210609103054027/Shenzhen Science and Technology Innovation Commission
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

