DEPDC1 as a metabolic target regulates glycolysis in renal cell carcinoma through AKT/mTOR/HIF1α pathway
- PMID: 39068164
- PMCID: PMC11283501
- DOI: 10.1038/s41419-024-06913-1
DEPDC1 as a metabolic target regulates glycolysis in renal cell carcinoma through AKT/mTOR/HIF1α pathway
Abstract
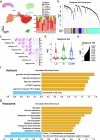
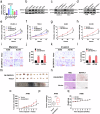
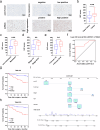
Renal cell carcinoma (RCC) is considered a "metabolic disease" characterized by elevated glycolysis in patients with advanced RCC. Tyrosine kinase inhibitor (TKI) therapy is currently an important treatment option for advanced RCC, but drug resistance may develop in some patients. Combining TKI with targeted metabolic therapy may provide a more effective approach for patients with advanced RCC. An analysis of 14 RCC patients (including three needle biopsy samples with TKI resistance) revealed by sing-cell RNA sequencing (scRNA-seq) that glycolysis played a crucial role in poor prognosis and drug resistance in RCC. TCGA-KIRC and glycolysis gene set analysis identified DEPDC1 as a target associated with malignant progression and drug resistance in KIRC. Subsequent experiments demonstrated that DEPDC1 promoted malignant progression and glycolysis of RCC, and knockdown DEPDC1 could reverse TKI resistance in RCC cell lines. Bulk RNA sequencing (RNA-seq) and non-targeted metabolomics sequencing suggested that DEPDC1 may regulate RCC glycolysis via AKT/mTOR/HIF1α pathway, a finding supported by protein-level analysis. Clinical tissue samples from 98 RCC patients demonstrated that DEPDC1 was associated with poor prognosis and predicted RCC metastasis. In conclusion, this multi-omics analysis suggests that DEPDC1 could serve as a novel target for TKI combined with targeted metabolic therapy in advanced RCC patients with TKI resistance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

