Glucocorticoid receptor controls atopic dermatitis inflammation via functional interactions with P63 and autocrine signaling in epidermal keratinocytes
- PMID: 39069531
- PMCID: PMC11284228
- DOI: 10.1038/s41419-024-06926-w
Glucocorticoid receptor controls atopic dermatitis inflammation via functional interactions with P63 and autocrine signaling in epidermal keratinocytes
Abstract
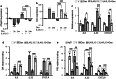
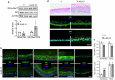
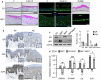
Atopic dermatitis (AD), a prevalent chronic inflammatory disease with multifactorial etiology, features epidermal barrier defects and immune overactivation. Synthetic glucocorticoids (GCs) are widely prescribed for treating AD due to their anti-inflammatory actions; however, mechanisms are incompletely understood. Defective local GC signaling due to decreased production of endogenous ligand and/or GC receptor (GR) levels was reported in prevalent inflammatory skin disorders; whether this is a consequence or contributing factor to AD pathology is unclear. To identify the chromatin-bound cell-type-specific GR protein interactome in keratinocytes, we used rapid immunoprecipitation of endogenous proteins and mass spectrometry identifying 145 interactors that increased upon dexamethasone treatment. GR-interacting proteins were enriched in p53/p63 signaling, including epidermal transcription factors with critical roles in AD pathology. Previous analyses indicating mirrored AD-like phenotypes between P63 overexpression and GR loss in epidermis, and our data show an intricate relationship between these transcription factors in human keratinocytes, identifying TP63 as a direct GR target. Dexamethasone treatment counteracted transcriptional up-regulation of inflammatory markers by IL4/IL13, known to mimic AD, causing opposite shifts in GR and P63 genomic binding. Indeed, IL4/IL13 decreased GR and increased P63 levels in cultured keratinocytes and human epidermal equivalents (HEE), consistent with GR down-regulation and increased P63 expression in AD lesions vs normal skin. Moreover, GR knockdown (GRKD) resulted in constitutive increases in P63, phospho-P38 and S100A9, IL6, and IL33. Also, GRKD culture supernatants showed increased autocrine production of TH2-/TH1-/TH17-TH22-associated factors including IL4, CXCL10, CXCL11, and CXCL8. GRKD HEEs showed AD-like features including hyperplasia and abnormal differentiation, resembling phenotypes observed with GR antagonist or IL4/IL13 treatment. The simultaneous GR/P63 knockdown partially reversed constitutive up-regulation of inflammatory genes in GRKD. In summary, our data support a causative role for GR loss in AD pathogenesis via functional interactions with P63 and autocrine signaling in epidermal keratinocytes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bieber T, Paller AS, Kabashima K, Feely M, Rueda MJ, Ross Terres JA, et al. Atopic dermatitis: pathomechanisms and lessons learned from novel systemic therapeutic options. J Eur Acad Dermatol Venereol. 2022;36:1432–49. https://pubmed.ncbi.nlm.nih.gov/35575442/. 10.1111/jdv.18225 - DOI - PubMed
-
- Meesters LD, Niehues H, Johnston L, Smits JPH, Zeeuwen PLJM, Brown SJ, et al. Keratinocyte signaling in atopic dermatitis: Investigations in organotypic skin models toward clinical application. J Allergy Clin Immunol. 2023;151:1231–5. https://pubmed.ncbi.nlm.nih.gov/36841264/. 10.1016/j.jaci.2023.02.012 - DOI - PubMed
-
- Strehl C, Ehlers L, Gaber T, Buttgereit F. Glucocorticoids-all-rounders tackling the versatile players of the immune system. Front Immunol. 2019;10:1744. https://pubmed.ncbi.nlm.nih.gov/31396235/. 10.3389/fimmu.2019.01744 - DOI - PMC - PubMed
-
- Vettorazzi S, Nalbantoglu D, Gebhardt JCM, Tuckermann J. A guide to changing paradigms of glucocorticoid receptor function-a model system for genome regulation and physiology. FEBS J. 2022;289:5718–43. https://pubmed.ncbi.nlm.nih.gov/34213830/. 10.1111/febs.16100 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

