Pathways to a Shiny Future: Building the Foundation for Computational Physical Chemistry and Biophysics in 2050
- PMID: 39069976
- PMCID: PMC11274290
- DOI: 10.1021/acsphyschemau.4c00003
Pathways to a Shiny Future: Building the Foundation for Computational Physical Chemistry and Biophysics in 2050
Abstract
In the last quarter-century, the field of molecular dynamics (MD) has undergone a remarkable transformation, propelled by substantial enhancements in software, hardware, and underlying methodologies. In this Perspective, we contemplate the future trajectory of MD simulations and their possible look at the year 2050. We spotlight the pivotal role of artificial intelligence (AI) in shaping the future of MD and the broader field of computational physical chemistry. We outline critical strategies and initiatives that are essential for the seamless integration of such technologies. Our discussion delves into topics like multiscale modeling, adept management of ever-increasing data deluge, the establishment of centralized simulation databases, and the autonomous refinement, cross-validation, and self-expansion of these repositories. The successful implementation of these advancements requires scientific transparency, a cautiously optimistic approach to interpreting AI-driven simulations and their analysis, and a mindset that prioritizes knowledge-motivated research alongside AI-enhanced big data exploration. While history reminds us that the trajectory of technological progress can be unpredictable, this Perspective offers guidance on preparedness and proactive measures, aiming to steer future advancements in the most beneficial and successful direction.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
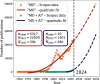
 , are shown in the inlet rectangles and
compared with publication data from the years 1998 and 2023 (N1998 and N2023,
respectively). The utilized Scopus data set is available on Zenodo
under DOI:
, are shown in the inlet rectangles and
compared with publication data from the years 1998 and 2023 (N1998 and N2023,
respectively). The utilized Scopus data set is available on Zenodo
under DOI: 
References
-
- Ciccotti G.; Dellago C.; Ferrario M.; Hernández E. R.; Tuckerman M. E. Molecular simulations: past, present, and future (a Topical Issue in EPJB). Eur. Phys. J. B 2022, 95, 1–12. 10.1140/epjb/s10051-021-00249-x. - DOI
-
- Frenkel D.; Smit B.. Underst. Mol. Simul. from Algorithms to Appl.. Third Ed.; Elsevier, 2023; pp 1–728, DOI: 10.1016/C2009-0-63921-0. - DOI
Publication types
LinkOut - more resources
Full Text Sources
