A genome-first approach to characterize DICER1 pathogenic variant prevalence, penetrance and cancer, thyroid, and other phenotypes in 2 population-scale cohorts
- PMID: 39070603
- PMCID: PMC11271802
- DOI: 10.1016/j.gimo.2024.101846
A genome-first approach to characterize DICER1 pathogenic variant prevalence, penetrance and cancer, thyroid, and other phenotypes in 2 population-scale cohorts
Abstract
Purpose: Population-scale, exome-sequenced cohorts with linked electronic health records (EHR) permit genome-first exploration of phenotype. Phenotype and cancer risk are well-characterized in children with a pathogenic DICER1 (HGNC ID:17098) variant. Here, the prevalence, penetrance and phenotype of pathogenic germline DICER1 variants in adults was investigated in two population-scale cohorts.
Methods: Variant pathogenicity was classified using published DICER1 ClinGen criteria in the UK Biobank (469,787 exomes; unrelated: 437,663) and Geisinger (170,503 exomes; unrelated: 109,789) cohorts. In the UK Biobank cohort, cancer diagnoses in the EHR, cancer and death registry were queried. For the Geisinger cohort, the Geisinger Cancer Registry and EHR were queried.
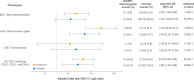
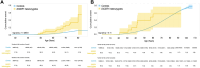
Results: In the UK Biobank, there were 46 unique pathogenic DICER1 variants in 57 individuals (1:8,242;95%CI:1:6,362-1:10,677). In Geisinger, there were 16 unique pathogenic DICER1 variants (including one microdeletion) in 21 individuals (1:8,119;95%CI:1:5,310-1:12,412). Cohorts were well-powered to find larger effect sizes for common cancers. Cancers were not significantly enriched in DICER1 heterozygotes; however, there was a ~4-fold increased risk for thyroid disease in both cohorts. There were multiple ICD10 codes enriched >2-fold in both cohorts.
Conclusion: Estimates of pathogenic germline DICER1 prevalence, thyroid disease penetrance and cancer phenotype from genomically ascertained adults are determined in two large cohorts.
Keywords: DICER1; DICER1 syndrome; healthcare population; penetrance; prevalence.
Conflict of interest statement
Conflict of Interest Statement The authors declare no conflicts of interest.
Figures

References
-
- Schultz K.A.P., Stewart D.R., Kamihara J., et al. In: GeneReviews®. Adam M.P., Mirzaa G.M., Pagon R.A., et al., editors. University of Washington; Seattle: 1993-2020. DICER1 tumor predisposition; pp. 1–34. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

