This is a preprint.
Epigenome-wide DNA Methylation Association Study of CHIP Provides Insight into Perturbed Gene Regulation
- PMID: 39070619
- PMCID: PMC11276001
- DOI: 10.21203/rs.3.rs-4656898/v1
Epigenome-wide DNA Methylation Association Study of CHIP Provides Insight into Perturbed Gene Regulation
Update in
-
Epigenome-wide DNA methylation association study of CHIP provides insight into perturbed gene regulation.Nat Commun. 2025 May 20;16(1):4678. doi: 10.1038/s41467-025-59333-w. Nat Commun. 2025. PMID: 40393957 Free PMC article.
Abstract
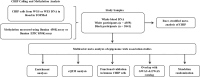
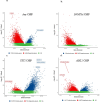
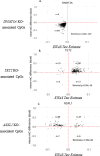
With age, hematopoietic stem cells can acquire somatic mutations in leukemogenic genes that confer a proliferative advantage in a phenomenon termed "clonal hematopoiesis of indeterminate potential" (CHIP). How these mutations confer a proliferative advantage and result in increased risk for numerous age-related diseases remains poorly understood. We conducted a multiracial meta-analysis of epigenome-wide association studies (EWAS) of CHIP and its subtypes in four cohorts (N=8196) to elucidate the molecular mechanisms underlying CHIP and illuminate how these changes influence cardiovascular disease risk. The EWAS findings were functionally validated using human hematopoietic stem cell (HSC) models of CHIP. A total of 9615 CpGs were associated with any CHIP, 5990 with DNMT3A CHIP, 5633 with TET2 CHIP, and 6078 with ASXL1 CHIP (P <1×10-7). CpGs associated with CHIP subtypes overlapped moderately, and the genome-wide DNA methylation directions of effect were opposite for TET2 and DNMT3A CHIP, consistent with their opposing effects on global DNA methylation. There was high directional concordance between the CpGs shared from the meta-EWAS and human edited CHIP HSCs. Expression quantitative trait methylation analysis further identified transcriptomic changes associated with CHIP-associated CpGs. Causal inference analyses revealed 261 CHIP-associated CpGs associated with cardiovascular traits and all-cause mortality (FDR adjusted p-value <0.05). Taken together, our study sheds light on the epigenetic changes impacted by CHIP and their associations with age-related disease outcomes. The novel genes and pathways linked to the epigenetic features of CHIP may serve as therapeutic targets for preventing or treating CHIP-mediated diseases.
Conflict of interest statement
Competing interests F.P. is an employee of Biomodal. L.M.R serves as a consultant for the NHLBI TOPMed Administrative Coordinating Center (through Westat). P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, TenSixteen Bio, and Tourmaline Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. Psaty serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. No other authors have competing interests. The Jimma University Internal Review Board approved the study ensuring all procedures involving human participation adhere to ethical guidelines established by both the Institution and National Research Committee before any research began.
Figures
Similar articles
-
Epigenome-wide DNA methylation association study of CHIP provides insight into perturbed gene regulation.Nat Commun. 2025 May 20;16(1):4678. doi: 10.1038/s41467-025-59333-w. Nat Commun. 2025. PMID: 40393957 Free PMC article.
-
Clonal hematopoiesis of indeterminate potential, DNA methylation, and risk for coronary artery disease.Nat Commun. 2022 Sep 12;13(1):5350. doi: 10.1038/s41467-022-33093-3. Nat Commun. 2022. PMID: 36097025 Free PMC article.
-
Incorporation of DNA methylation quantitative trait loci (mQTLs) in epigenome-wide association analysis: application to birthweight effects in neonatal whole blood.Clin Epigenetics. 2022 Dec 1;14(1):158. doi: 10.1186/s13148-022-01385-6. Clin Epigenetics. 2022. PMID: 36457128 Free PMC article.
-
[Aging and clonal hematopoesis.].Adv Gerontol. 2024;37(3):266-275. Adv Gerontol. 2024. PMID: 39139119 Review. Russian.
-
Clonal hematopoiesis driven by DNMT3A and TET2 mutations: role in monocyte and macrophage biology and atherosclerotic cardiovascular disease.Curr Opin Hematol. 2022 Jan 1;29(1):1-7. doi: 10.1097/MOH.0000000000000688. Curr Opin Hematol. 2022. PMID: 34654019 Free PMC article. Review.
References
-
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98. Epub 20141126. doi: 10.1056/NEJMoa1408617. - DOI - PMC - PubMed
-
- Miller PG, Qiao D, Rojas-Quintero J, Honigberg MC, Sperling AS, Gibson CJ, Bick AG, Niroula A, McConkey ME, Sandoval B, Miller BC, Shi W, Viswanathan K, Leventhal M, Werner L, Moll M, Cade BE, Barr RG, Correa A, Cupples LA, Gharib SA, Jain D, Gogarten SM, Lange LA, London SJ, Manichaikul A, O’Connor GT, Oelsner EC, Redline S, Rich SS, Rotter JI, Ramachandran V, Yu B, Sholl L, Neuberg D, Jaiswal S, Levy BD, Owen CA, Natarajan P, Silverman EK, van Galen P, Tesfaigzi Y, Cho MH, Ebert BL, Copdgene Study Investigators NHL, Blood Institute Trans-Omics for Precision Medicine C. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood. 2022;139(3):357–68. doi: 10.1182/blood.2021013531. - DOI - PMC - PubMed
Publication types
Grants and funding
- HHSN268201100037C/HL/NHLBI NIH HHS/United States
- HHSN268201800012C/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- HHSN268201800014C/HL/NHLBI NIH HHS/United States
- HHSN268201700002C/HL/NHLBI NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- HHSN268201800011C/HL/NHLBI NIH HHS/United States
- HHSN268201800015I/HB/NHLBI NIH HHS/United States
- HHSN268201700004C/HL/NHLBI NIH HHS/United States
- HHSN268201800011I/HB/NHLBI NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- R01 HL103612/HL/NHLBI NIH HHS/United States
- N01 HC085080/HL/NHLBI NIH HHS/United States
- T32 GM080178/GM/NIGMS NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- HHSN268201500001C/HL/NHLBI NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- HHSN268201800014I/HB/NHLBI NIH HHS/United States
- HHSN268200800007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- R01 HL168894/HL/NHLBI NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- HHSN268201700001I/HL/NHLBI NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- HHSN268201800013I/MD/NIMHD NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201700004I/HL/NHLBI NIH HHS/United States
- HHSN268201500001I/HL/NHLBI NIH HHS/United States
- HHSN268201800012I/HL/NHLBI NIH HHS/United States
- 75N92021D00006/HL/NHLBI NIH HHS/United States
- HHSN268201700005C/HL/NHLBI NIH HHS/United States
- HHSN268201700001C/HL/NHLBI NIH HHS/United States
- HHSN268201700003C/HL/NHLBI NIH HHS/United States
- HHSN268201700002I/HL/NHLBI NIH HHS/United States
- HHSN268201800010I/HB/NHLBI NIH HHS/United States
- HHSN268201700005I/HL/NHLBI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- HHSN268201700003I/HL/NHLBI NIH HHS/United States
- R01 HL148050/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources

