This is a preprint.
Massive experimental quantification of amyloid nucleation allows interpretable deep learning of protein aggregation
- PMID: 39071305
- PMCID: PMC11275847
- DOI: 10.1101/2024.07.13.603366
Massive experimental quantification of amyloid nucleation allows interpretable deep learning of protein aggregation
Update in
-
Massive experimental quantification allows interpretable deep learning of protein aggregation.Sci Adv. 2025 May 2;11(18):eadt5111. doi: 10.1126/sciadv.adt5111. Epub 2025 Apr 30. Sci Adv. 2025. PMID: 40305601 Free PMC article.
Abstract
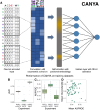
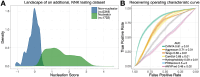
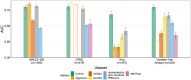
Protein aggregation is a pathological hallmark of more than fifty human diseases and a major problem for biotechnology. Methods have been proposed to predict aggregation from sequence, but these have been trained and evaluated on small and biased experimental datasets. Here we directly address this data shortage by experimentally quantifying the amyloid nucleation of >100,000 protein sequences. This unprecedented dataset reveals the limited performance of existing computational methods and allows us to train CANYA, a convolution-attention hybrid neural network that accurately predicts amyloid nucleation from sequence. We adapt genomic neural network interpretability analyses to reveal CANYA's decision-making process and learned grammar. Our results illustrate the power of massive experimental analysis of random sequence-spaces and provide an interpretable and robust neural network model to predict amyloid nucleation.
Conflict of interest statement
Competing interests The authors have declared no competing interests.
Figures




References
-
- Chiti F. & Dobson C. M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 86, 27–68 (2017). - PubMed
-
- Fowler D. M., Koulov A. V., Balch W. E. & Kelly J. W. Functional amyloid--from bacteria to humans. Trends Biochem. Sci. 32, 217–224 (2007). - PubMed
-
- Shire S. J. Formulation and manufacturability of biologics. Curr. Opin. Biotechnol. 20, 708–714 (2009). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
