This is a preprint.
Proxi-RIMS-seq2 applied to native microbiomes uncovers hundreds of known and novel m5C methyltransferase specificities
- PMID: 39071437
- PMCID: PMC11275837
- DOI: 10.1101/2024.07.15.603628
Proxi-RIMS-seq2 applied to native microbiomes uncovers hundreds of known and novel m5C methyltransferase specificities
Update in
-
Proxi-RIMS-seq2 applied to native microbiomes uncovers hundreds of known and novel m5C methyltransferase specificities.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf226. doi: 10.1093/nar/gkaf226. Nucleic Acids Res. 2025. PMID: 40156868 Free PMC article.
Abstract
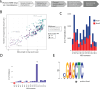
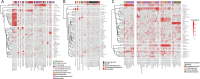
Methylation patterns in bacteria can be used to study Restriction-Modification (RM) or other defense systems with novel properties. While m4C and m6A methylation is well characterized mainly through PacBio sequencing, the landscape of m5C methylation is under-characterized. To bridge this gap, we performed RIMS-seq2 on microbiomes composed of resolved assemblies of distinct genomes through proximity ligation. This high-throughput approach enables the identification of m5C methylated motifs and links them to cognate methyltransferases directly on native microbiomes without the need to isolate bacterial strains. Methylation patterns can also be identified on viral DNA and compared to host DNA, strengthening evidence for virus-host interaction. Applied to three different microbiomes, the method unveils over 1900 motifs that were deposited in REBASE. The motifs include a novel 8-base recognition site (CATm5CGATG) that was experimentally validated by characterizing its cognate methyltransferase. Our findings suggest that microbiomes harbor arrays of untapped m5C methyltransferase specificities, providing insights to bacterial biology and biotechnological applications.
Conflict of interest statement
WY, YL, RR and LE are employees of New England Biolabs, Inc. a manufacturer of restriction enzymes and molecular biology reagents. ER, HM, ZS, BA, and IL are employees of Phase Genomics, the developer of metagenomic proximity ligation technology.
Figures

References
-
- Anton Brian P., and Roberts Richard J.. 2021. “Beyond Restriction Modification: Epigenomic Roles of DNA Methylation in Prokaryotes.” Annual Review of Microbiology 75 (October): 129–49. - PubMed
-
- Baum Chloé, Lin Yu-Cheng, Fomenkov Alexey, Anton Brian P., Chen Lixin, Yan Bo, Evans Thomas C., Roberts Richard J., Tolonen Andrew C., and Ettwiller Laurence. 2021. “Rapid Identification of Methylase Specificity (RIMS-Seq) Jointly Identifies Methylated Motifs and Generates Shotgun Sequencing of Bacterial Genomes.” Nucleic Acids Research 49 (19): e113. - PMC - PubMed
-
- Bickhart Derek M., Kolmogorov Mikhail, Tseng Elizabeth, Portik Daniel M., Korobeynikov Anton, Tolstoganov Ivan, Uritskiy Gherman, et al. 2022. “Generating Lineage-Resolved, Complete Metagenome-Assembled Genomes from Complex Microbial Communities.” Nature Biotechnology 40 (5): 711–19. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
