This is a preprint.
"Off-pore" nucleoporins relocalize heterochromatic breaks through phase separation
- PMID: 39071440
- PMCID: PMC11275802
- DOI: 10.1101/2023.12.07.570729
"Off-pore" nucleoporins relocalize heterochromatic breaks through phase separation
Abstract
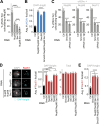
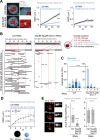
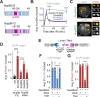
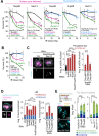
Phase separation forms membraneless compartments in the nuclei, including by establishing heterochromatin "domains" and repair foci. Pericentromeric heterochromatin mostly comprises repeated sequences prone to aberrant recombination, and "safe" homologous recombination (HR) repair of these sequences requires the movement of repair sites to the nuclear periphery before Rad51 recruitment and strand invasion. How this mobilization initiates is unknown, and the contribution of phase separation to these dynamics is unclear. Here, we show that Nup98 nucleoporin is recruited to heterochromatic repair sites before relocalization through Sec13 or Nup88 nucleoporins, and downstream from the Smc5/6 complex and SUMOylation. Remarkably, the phase separation properties of Nup98 are required and sufficient to mobilize repair sites and exclude Rad51, thus preventing aberrant recombination while promoting HR repair. Disrupting this pathway results in heterochromatin repair defects and widespread chromosome rearrangements, revealing a novel "off-pore" role for nucleoporins and phase separation in nuclear dynamics and genome integrity in a multicellular eukaryote.
Keywords: Heterochromatin repair; Nup88; Nup98; Sec13; condensates; double-strand break repair; homologous recombination; nuclear architecture; phase separation; repair dynamics.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



References
-
- Peng J. C. & Karpen G. H. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev 18, 204–211 (2008). https://doi.org:S0959-437X(08)00020-8 [pii] 10.1016/j.gde.2008.01.021 - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
