This is a preprint.
Emergence of cellular nematic order is a conserved feature of gastrulation in animal embryos
- PMID: 39071444
- PMCID: PMC11275887
- DOI: 10.1101/2024.07.11.603175
Emergence of cellular nematic order is a conserved feature of gastrulation in animal embryos
Update in
-
Emergence of cellular nematic order is a conserved feature of gastrulation in animal embryos.Nat Commun. 2025 Jul 1;16(1):5946. doi: 10.1038/s41467-025-61045-0. Nat Commun. 2025. PMID: 40595575 Free PMC article.
Abstract
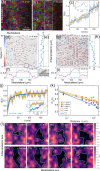
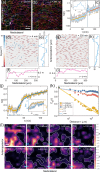
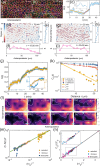
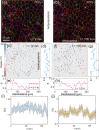
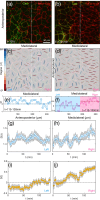
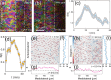
Cells undergo dramatic morphological changes during embryogenesis, yet how these changes affect the formation of ordered tissues remains elusive. Here, we show that a phase transition leading to the formation of a nematic liquid crystal state during gastrulation in the development of embryos of fish, frogs, and fruit flies occurs by a common mechanism despite substantial differences between these evolutionarily distant animals. Importantly, nematic order forms early before any discernible changes in the shapes of cells. All three species exhibit similar propagation of the nematic phase, reminiscent of nucleation and growth mechanisms. The spatial correlations in the nematic phase in the notochord region are long-ranged and follow a similar power-law decay ( ) with less than unity, indicating a common underlying physical mechanism. To explain the common physical mechanism, we created a theoretical model that not only explains the experimental observations but also predicts that the nematic phase should be disrupted upon loss of planar cell polarity (frog), cell adhesion (frog), and notochord boundary formation (zebrafish). Gene knockdown or mutational studies confirm the theoretical predictions. The combination of experiments and theory provides a unified framework for understanding the potentially universal features of metazoan embryogenesis, in the process shedding light on the advent of ordered structures during animal development.
Conflict of interest statement
Competing Interests: No competing interests declared.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
