Progesterone modulates cell growth via integrin αvβ3-dependent pathway in progesterone receptor-negative MDA-MB-231 cells
- PMID: 39071644
- PMCID: PMC11283053
- DOI: 10.1016/j.heliyon.2024.e34006
Progesterone modulates cell growth via integrin αvβ3-dependent pathway in progesterone receptor-negative MDA-MB-231 cells
Abstract
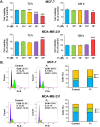
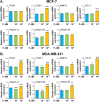
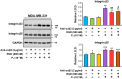
Progesterone (P4) plays a pivotal role in regulating the cancer progression of various types, including breast cancer, primarily through its interaction with the P4 receptor (PR). In PR-negative breast cancer cells, P4 appears to function in mediating cancer progression, such as cell growth. However, the mechanisms underlying the roles of P4 in PR-negative breast cancer cells remain incompletely understood. This study aimed to investigate the effects of P4 on cell proliferation, gene expression, and signal transduction in PR-negative MDA-MB-231 breast cancer cells. P4-activated genes, associated with proliferation in breast cancer cells, exhibit a stimulating effect on cell growth in PR-negative MDA-MB-231 cells, while demonstrating an inhibitory impact in PR-positive MCF-7 cells. The use of arginine-glycine-aspartate (RGD) peptide successfully blocked P4-induced extracellular signal-regulated kinase 1/2 (ERK1/2) activation, aligning with computational models of P4 binding to integrin αvβ3. Disrupting integrin αvβ3 binding with RGD peptide or anti-integrin αvβ3 antibody altered P4-induced expression of proliferative genes and modified P4-induced cell growth in breast cancer cells. In conclusion, integrin αvβ3 appears to mediate P4-induced ERK1/2 signal pathway to regulate proliferation via alteration of proliferation-related gene expression in PR-negative breast cancer cells.
Keywords: Breast cancer; Integrin αvβ3; Progesterone; Progesterone receptor; Proliferation.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Ferlay J E.M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F., Global Cancer Observatory: Cancer Today (version 1.1). Lyon, France: International Agency for Research on Cancer (2020). https://gco.iarc.who.int/todayhttps://gco.iarc.fr/today. (Accessed 7 September 2020).
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

