MiR-30c suppresses the proliferation, metastasis and polarity reversal of tumor cell clusters by targeting MTDH in invasive micropapillary carcinoma of the breast
- PMID: 39071710
- PMCID: PMC11279262
- DOI: 10.1016/j.heliyon.2024.e33938
MiR-30c suppresses the proliferation, metastasis and polarity reversal of tumor cell clusters by targeting MTDH in invasive micropapillary carcinoma of the breast
Abstract
Purpose: Invasive micropapillary carcinoma (IMPC) of the breast has a high propensity for lymphovascular invasion and axillary lymph node metastasis and displays an 'inside-out' growth pattern, but the molecular mechanism of invasion, metastasis and cell polarity reversal in IMPC is unclear.
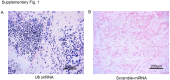
Methods: and Patients: Cell growth curves, tumor sphere formation assays, transwell assays, mouse xenograft model and immunofluorescence were evaluated to investigate the effects of miR-30c and MTDH. Dual luciferase reporter assays was performed to confirm that the MTDH (metadherin) 3'UTR bound to miR-30c. MiRNA in situ hybridization (ISH) and immunohistochemistry (IHC) were carried out on IMPC patient tissues for miR-30c and MTDH expression, respectively.
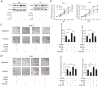
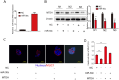
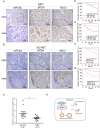
Results: We found miR-30c as a tumor suppressor gene in cell proliferation, metastasis and polarity reversal of IMPC. Overexpression of miR-30c inhibited cell growth and metastasis in vitro and in vivo. MiR-30c could directly target the MTDH 3'UTR. MiR-30c overexpression inhibited breast cancer cell proliferation, invasion and metastasis by targeting MTDH. Moreover, miR-30c/MTDH axis could also regulate cell polarity reversal of IMPC. By ISH and IHC analyses, miR-30c and MTDH were significantly correlated with tumor size, lymph nodule status and tumor grade, the 'inside-out' growth pattern, overall survival (OS) and disease-free survival (DFS) in IMPC patients.
Conclusions: Overall, miR-30c/MTDH axis was responsible for tumor proliferation, metastasis and polarity reversal. It may provide promising therapeutic targets and prognostic biomarkers for patients with IMPC.
Keywords: Breast cancer; Invasive micropapillary carcinoma (IMPC); Metastasis; MiRNA; Polarity reversal; Proliferation.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Lifu reports was provided by none. Lifu reports a relationship with none that includes:. Lifu has patent none pending to none. none If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Overexpression of β1 integrin contributes to polarity reversal and a poor prognosis of breast invasive micropapillary carcinoma.Oncotarget. 2017 Nov 30;9(4):4338-4353. doi: 10.18632/oncotarget.22774. eCollection 2018 Jan 12. Oncotarget. 2017. PMID: 29435106 Free PMC article.
-
MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin.Oncogene. 2014 Jun 12;33(24):3119-28. doi: 10.1038/onc.2013.286. Epub 2013 Jul 15. Oncogene. 2014. PMID: 23851509
-
MiR-524 suppressed the progression of oral squamous cell carcinoma by suppressing Metadherin and NF-κB signaling pathway in OSCC cell lines.Arch Oral Biol. 2021 May;125:105090. doi: 10.1016/j.archoralbio.2021.105090. Epub 2021 Feb 22. Arch Oral Biol. 2021. PMID: 33676362
-
Invasive micropapillary carcinoma: a distinct type of adenocarcinomas in the gastrointestinal tract.World J Gastroenterol. 2014 Apr 28;20(16):4597-606. doi: 10.3748/wjg.v20.i16.4597. World J Gastroenterol. 2014. PMID: 24782612 Free PMC article. Review.
-
An overview of invasive micropapillary carcinoma of the breast: past, present, and future.Front Oncol. 2024 Nov 15;14:1435421. doi: 10.3389/fonc.2024.1435421. eCollection 2024. Front Oncol. 2024. PMID: 39619442 Free PMC article. Review.
Cited by
-
Transmucosal Delivery of miR-30c-5p by Chitosan Nanoparticles for Oral Squamous Cell Carcinoma.Int J Nanomedicine. 2025 Jul 19;20:9179-9194. doi: 10.2147/IJN.S524558. eCollection 2025. Int J Nanomedicine. 2025. PMID: 40703474 Free PMC article.
References
-
- Siriaunkgul S., Tavassoli F.A. Invasive micropapillary carcinoma of the breast. Mod. Pathol. 1993;6:660–662. - PubMed
-
- WHO Classification of Tumors Editorial Board . Breast Tumors. fifth ed. IARC; Lyon: 2019. WHO classification of tumors.
-
- Ohtsuki Y., Kuroda N., Yunoki S., Murakami S., Mizukami Y., Okada Y., Iguchi M., Lee G.H., Furihata M. Immunohistochemical analysis of invasive micropapillary carcinoma pattern in four cases of gastric cancer. Med. Mol. Morphol. 2013;46:114–121. - PubMed
-
- Verdu M., Roman R., Calvo M., Rodon N., Garcia B., Gonzalez M., Vidal A., Puig X. Clinicopathological and molecular characterization of colorectal micropapillary carcinoma. Mod. Pathol. 2011;24:729–738. - PubMed
-
- Koga K., Hamasaki M., Kato F., Aoki M., Hayashi H., Iwasaki A., Kataoka H., Nabeshima K. Association of c-Met phosphorylation with micropapillary pattern and small cluster invasion in pT1-size lung adenocarcinoma. Lung Cancer. 2013;82:413–419. - PubMed
LinkOut - more resources
Full Text Sources

