An evaluation of nucleic acid-based molecular methods for the detection of plant viruses: a systematic review
- PMID: 39071869
- PMCID: PMC11269559
- DOI: 10.1007/s13337-024-00863-0
An evaluation of nucleic acid-based molecular methods for the detection of plant viruses: a systematic review
Abstract
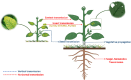
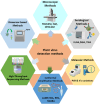
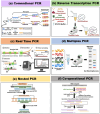
Precise and timely diagnosis of plant viruses is a prerequisite for the implementation of efficient management strategies, considering factors like globalization of trade and climate change facilitating the spread of viruses that lead to agriculture yield losses of billions yearly worldwide. Symptomatic diagnosis alone may not be reliable due to the diverse symptoms and confusion with plant abiotic stresses. It is crucial to detect plant viruses accurately and reliably and do so with little time. A complete understanding of the various detection methods is necessary to achieve this. Enzyme-linked immunosorbent assay (ELISA), has become more popular as a method for detecting viruses but faces limitations such as antibody availability, cost, sample volume, and time. Advanced techniques like polymerase chain reaction (PCR) have surpassed ELISA with its various sensitive variants. Over the last decade, nucleic acid-based molecular methods have gained popularity and have quickly replaced other techniques, such as serological techniques for detecting plant viruses due to their specificity and accuracy. Hence, this review enables the reader to understand the strengths and weaknesses of each molecular technique starting with PCR and its variations, along with various isothermal amplification followed by DNA microarrays, and next-generation sequencing (NGS). As a result of the development of new technologies, NGS is becoming more and more accessible and cheaper, and it looks possible that this approach will replace others as a favoured approach for carrying out regular diagnosis. NGS is also becoming the method of choice for identifying novel viruses.
Supplementary information: The online version contains supplementary material available at 10.1007/s13337-024-00863-0.
Keywords: Isothermal amplification; Molecular techniques; PCR; Plant virus detection; Virus diagnosis.
© The Author(s), under exclusive licence to Indian Virological Society 2024. Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestsThe authors declare that they have no conflict of interest.
Figures
Similar articles
-
Methods in virus diagnostics: from ELISA to next generation sequencing.Virus Res. 2014 Jun 24;186:20-31. doi: 10.1016/j.virusres.2013.12.007. Epub 2013 Dec 19. Virus Res. 2014. PMID: 24361981 Review.
-
Recent Advances on the Multiplex Molecular Detection of Plant Viruses and Viroids.Front Microbiol. 2018 Sep 10;9:2087. doi: 10.3389/fmicb.2018.02087. eCollection 2018. Front Microbiol. 2018. PMID: 30250456 Free PMC article. Review.
-
Detection of Plant Viruses and Disease Management: Relevance of Genetic Diversity and Evolution.Front Plant Sci. 2020 Jul 17;11:1092. doi: 10.3389/fpls.2020.01092. eCollection 2020. Front Plant Sci. 2020. PMID: 32765569 Free PMC article. Review.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
-
FnCas9-based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV-2 variants on a paper strip.Elife. 2021 Jun 9;10:e67130. doi: 10.7554/eLife.67130. Elife. 2021. PMID: 34106048 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

