This is a preprint.
Ex vivo susceptibility to antimalarial drugs and polymorphisms in drug resistance genes of African Plasmodium falciparum, 2016-2023: a genotype-phenotype association study
- PMID: 39072017
- PMCID: PMC11275679
- DOI: 10.1101/2024.07.17.24310448
Ex vivo susceptibility to antimalarial drugs and polymorphisms in drug resistance genes of African Plasmodium falciparum, 2016-2023: a genotype-phenotype association study
Update in
-
Assessment of ex vivo antimalarial drug efficacy in African Plasmodium falciparum parasite isolates, 2016-2023: a genotype-phenotype association study.EBioMedicine. 2025 Aug;118:105835. doi: 10.1016/j.ebiom.2025.105835. Epub 2025 Jul 9. EBioMedicine. 2025. PMID: 40639048 Free PMC article.
Abstract
Background: Given the altered responses to both artemisinins and lumefantrine in Eastern Africa, monitoring antimalarial drug resistance in all African countries is paramount.
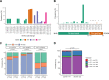
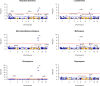
Methods: We measured the susceptibility to six antimalarials using ex vivo growth inhibition assays (IC50) for a total of 805 Plasmodium falciparum isolates obtained from travelers returning to France (2016-2023), mainly from West and Central Africa. Isolates were sequenced using molecular inversion probes (MIPs) targeting fourteen drug resistance genes across the parasite genome.
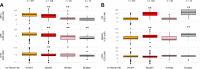
Findings: Ex vivo susceptibility to several drugs has significantly decreased in 2019-2023 versus 2016-2018 parasite samples: lumefantrine (median IC50: 23·0 nM [IQR: 14·4-35·1] in 2019-2023 versus 13·9 nM [8·42-21·7] in 2016-2018, p<0·0001), monodesethylamodiaquine (35·4 [21·2-51·1] versus 20·3 nM [15·4-33·1], p<0·0001), and marginally piperaquine (20·5 [16·5-26·2] versus 18.0 [14·2-22·4] nM, p<0·0001). Only four isolates carried a validated pfkelch13 mutation. Multiple mutations in pfcrt and one in pfmdr1 (N86Y) were significantly associated with altered susceptibility to multiple drugs. The susceptibility to lumefantrine was altered by pfcrt and pfmdr1 mutations in an additive manner, with the wild-type haplotype (pfcrt K76-pfmdr1 N86) exhibiting the least susceptibility.
Interpretation: Our study on P. falciparum isolates from West and Central Africa indicates a low prevalence of molecular markers of artemisinin resistance but a significant decrease in susceptibility to the partner drugs that have been the most widely used since a decade -lumefantrine and amodiaquine. These phenotypic changes likely mark parasite adaptation to sustained drug pressure and call for intensifying the monitoring of antimalarial drug resistance in Africa.
Funding: This work was supported by the French Ministry of Health (grant to the French National Malaria Reference Center) and by the Agence Nationale de la Recherche (ANR-17-CE15-0013-03 to JC). JAB was supported by NIH R01AI139520. JR postdoctoral fellowship was funded by Institut de Recherche pour le Développement.
Keywords: Antimalarial drug resistance; IC50 assay; ex-vivo susceptibility; genotype.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interest.
Figures


References
-
- Conrad MD, Rosenthal PJ. Antimalarial drug resistance in Africa: the calm before the storm? Lancet Infect Dis 2019; 19: e338–51. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
