This is a preprint.
Clinical Relevance of Computationally Derived Tubular Features: Spatial Relationships and the Development of Tubulointerstitial Scarring in MCD/FSGS
- PMID: 39072032
- PMCID: PMC11275675
- DOI: 10.1101/2024.07.19.24310619
Clinical Relevance of Computationally Derived Tubular Features: Spatial Relationships and the Development of Tubulointerstitial Scarring in MCD/FSGS
Update in
-
Clinical relevance of computationally derived tubular features and their spatial relationships with the interstitial microenvironment in minimal change disease/focal segmental glomerulosclerosis.Kidney Int. 2025 Aug;108(2):293-309. doi: 10.1016/j.kint.2025.04.026. Epub 2025 May 21. Kidney Int. 2025. PMID: 40409668
Abstract
Background: Visual scoring of tubular damage has limitations in capturing the full spectrum of structural changes and prognostic potential. We investigate if computationally quantified tubular features can enhance prognostication and reveal spatial relationships with interstitial fibrosis.
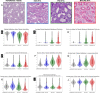
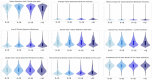
Methods: Deep-learning and image-processing-based segmentations were employed in N=254/266 PAS-WSIs from the NEPTUNE/CureGN datasets (135/153 focal segmental glomerulosclerosis and 119/113 minimal change disease) for: cortex, tubular lumen (TL), epithelium (TE), nuclei (TN), and basement membrane (TBM). N=104 pathomic features were extracted from these segmented tubular substructures and summarized at the patient level using summary statistics. The tubular features were quantified across the biopsy and in manually segmented regions of mature interstitial fibrosis and tubular atrophy (IFTA), pre-IFTA and non-IFTA in the NEPTUNE dataset. Minimum Redundancy Maximum Relevance was used in the NEPTUNE dataset to select features most associated with disease progression and proteinuria remission. Ridge-penalized Cox models evaluated their predictive discrimination compared to clinical/demographic data and visual-assessment. Models were evaluated in the CureGN dataset.
Results: N=9 features were predictive of disease progression and/or proteinuria remission. Models with tubular features had high prognostic accuracy in both NEPTUNE and CureGN datasets and increased prognostic accuracy for both outcomes (5.6%-7.7% and 1.6%-4.6% increase for disease progression and proteinuria remission, respectively) compared to conventional parameters alone in the NEPTUNE dataset. TBM thickness/area and TE simplification progressively increased from non- to pre- and mature IFTA.
Conclusions: Previously under-recognized, quantifiable, and clinically relevant tubular features in the kidney parenchyma can enhance understanding of mechanisms of disease progression and risk stratification.
Conflict of interest statement
Competing Interest Statement All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: FF, JJ and AJ have received financial support from NIH funding list in the acknowledgement. JZ has received financial support from NIDDK and NCATS for the submitted work and received grants from Boehringer-Ingelheim, Travere Therapeutics, Reliant Glycosciences, HiBio, and Takeda Pharmaceuticals in the past 3 years. JZ has also received an honorarium for technical expert panel participation from Booz Allen Hamilton. LM has received financial support from NIDDK and NCATS for the submitted work and received grants from Boehringer-Ingelheim, Travere Therapeutics, Reliant Glycosciences, HiBio and Takeda Pharmaceuticals. LM has also received consulting fee from Novartis, Calliditas and Travere and payment for educational events from WebMD/Medscape and MedLive/PlatformQ. JR has received grants from National Science Foundation Graduate Research Fellowship. LBH has received grants from NIDDK CureGN-Penn PCC, NIDDK Nephrotic Syndrome Rare Disease Clinical Research Network III and NIDDK Computational Pathology for Proteinuric Glomerulopathies. Additionally, LBH holds a leadership role in the Scientific Advisory Board of NephCure Kidney International. JH has received grants from NIH and Department of Defense. AM is an equity holder in Picture Health, Elucid Bioimaging, and Inspirata Inc. Currently he serves on the advisory board of Picture Health, and SimBioSys. AM currently consults for Takeda Inc. AM also has sponsored research agreements with AstraZeneca and Bristol Myers-Squibb. His technology has been licensed to Picture Health and Elucid Bioimaging. AM is also involved in 2 different R01 grants with Inspirata Inc. AM also serves as a member for the Frederick National Laboratory Advisory Committee. LB has received grants from NIH fundings listed in Acknowledgment, Nephcure and Haller Foundation. LB has also participated on a Data Safety Monitoring Board or Advisory Board for Vertex and holds a leadership role in the International Society of Glomerular Diseases.
Figures



References
-
- Morphometric and visual evaluation of fibrosis in renal biopsies - PubMed. Accessed May 30, 2024. https://pubmed.ncbi.nlm.nih.gov/21115619/ - PMC - PubMed
Publication types
Grants and funding
- U24 DK100845/DK/NIDDK NIH HHS/United States
- U01 DK100867/DK/NIDDK NIH HHS/United States
- U2C TR002818/TR/NCATS NIH HHS/United States
- R01 CA216579/CA/NCI NIH HHS/United States
- R01 LM013864/LM/NLM NIH HHS/United States
- U01 DK133090/DK/NIDDK NIH HHS/United States
- U54 DK083912/DK/NIDDK NIH HHS/United States
- C06 RR012463/RR/NCRR NIH HHS/United States
- UM1 DK100846/DK/NIDDK NIH HHS/United States
- U01 DK100876/DK/NIDDK NIH HHS/United States
- R43 EB028736/EB/NIBIB NIH HHS/United States
- UM1 DK100845/DK/NIDDK NIH HHS/United States
- UM1 DK100866/DK/NIDDK NIH HHS/United States
- R01 CA249992/CA/NCI NIH HHS/United States
- R01 CA220581/CA/NCI NIH HHS/United States
- R01 DK118431/DK/NIDDK NIH HHS/United States
- U01 DK100866/DK/NIDDK NIH HHS/United States
- U01 DK100846/DK/NIDDK NIH HHS/United States
- UM1 DK100876/DK/NIDDK NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- R01 CA208236/CA/NCI NIH HHS/United States
- U01 CA248226/CA/NCI NIH HHS/United States
- I01 BX004121/BX/BLRD VA/United States
- R01 CA257612/CA/NCI NIH HHS/United States
- U54 CA254566/CA/NCI NIH HHS/United States
- UM1 DK100867/DK/NIDDK NIH HHS/United States
- U01 CA239055/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
