Endotypes of severe neutrophilic and eosinophilic asthma from multi-omics integration of U-BIOPRED sputum samples
- PMID: 39073027
- PMCID: PMC11283589
- DOI: 10.1002/ctm2.1771
Endotypes of severe neutrophilic and eosinophilic asthma from multi-omics integration of U-BIOPRED sputum samples
Abstract
Background: Clustering approaches using single omics platforms are increasingly used to characterise molecular phenotypes of eosinophilic and neutrophilic asthma. Effective integration of multi-omics platforms should lead towards greater refinement of asthma endotypes across molecular dimensions and indicate key targets for intervention or biomarker development.
Objectives: To determine whether multi-omics integration of sputum leads to improved granularity of the molecular classification of severe asthma.
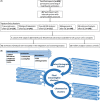
Methods: We analyzed six -omics data blocks-microarray transcriptomics, gene set variation analysis of microarray transcriptomics, SomaSCAN proteomics assay, shotgun proteomics, 16S microbiome sequencing, and shotgun metagenomic sequencing-from induced sputum samples of 57 severe asthma patients, 15 mild-moderate asthma patients, and 13 healthy volunteers in the U-BIOPRED European cohort. We used Monti consensus clustering algorithm for aggregation of clustering results and Similarity Network Fusion to integrate the 6 multi-omics datasets of the 72 asthmatics.
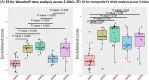
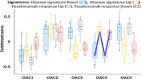
Results: Five stable omics-associated clusters were identified (OACs). OAC1 had the best lung function with the least number of severe asthmatics with sputum paucigranulocytic inflammation. OAC5 also had fewer severe asthma patients but the highest incidence of atopy and allergic rhinitis, with paucigranulocytic inflammation. OAC3 comprised only severe asthmatics with the highest sputum eosinophilia. OAC2 had the highest sputum neutrophilia followed by OAC4 with both clusters consisting of mostly severe asthma but with more ex/current smokers in OAC4. Compared to OAC4, there was higher incidence of nasal polyps, allergic rhinitis, and eczema in OAC2. OAC2 had microbial dysbiosis with abundant Moraxella catarrhalis and Haemophilus influenzae. OAC4 was associated with pathways linked to IL-22 cytokine activation, with the prediction of therapeutic response to anti-IL22 antibody therapy.
Conclusion: Multi-omics analysis of sputum in asthma has defined with greater granularity the asthma endotypes linked to neutrophilic and eosinophilic inflammation. Modelling diverse types of high-dimensional interactions will contribute to a more comprehensive understanding of complex endotypes.
Key points: Unsupervised clustering on sputum multi-omics of asthma subjects identified 3 out of 5 clusters with predominantly severe asthma. One severe asthma cluster was linked to type 2 inflammation and sputum eosinophilia while the other 2 clusters to sputum neutrophilia. One severe neutrophilic asthma cluster was linked to Moraxella catarrhalis and to a lesser extent Haemophilus influenzae while the second cluster to activation of IL-22.
Keywords: asthma endotype; consensus clustering; eosinophilic inflammation; gene set variation analysis; neutrophilic inflammation; pathogenic bacteria; severe asthma; similarity network fusion.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
Mrs. Zounemat‐Kermani has nothing to declare. Dr. Maitland‐van der Zee has received grants from Health Holland and she is the PI of a P4O2 (Precision Medicine for more Oxygen) public–private partnership sponsored by Health Holland involving many private partners that contribute in cash and/or in kind (Boehringer Ingelheim, Breathomix, Fluidda, Ortec Logiqcare, Philips, Quantib‐U, Smartfish, SODAQ, Thirona, TopMD and Novartis), received unrestricted research grants from GSK, Boehringer Ingelheim and Vertex, received consulting fees paid to her institution from Boehringer Ingelheim and AstraZeneca, and received honoraria for lectures paid to her institution from GlaxoSmithKline – outside the submitted work. Dr. Dahlén reports personal fees from AZ, Cayman Chemicals, GSK, Novartis, Regeneron, Sanofi, TEVA, outside the submitted work. Dr. Chung has received honoraria for participating in Advisory Board meetings of Roche, Merck, Shionogi and Rickett‐Beckinson and has also been renumerated for speaking engagements for Novartis and AZ. Dr. Djukanovic declares consulting fees from Synairgen, Sanofi and Galapagos, lecture fees from GSK, AZ and Airways Vista and he holds shares from Synairgen. Dr. Li, Mr. Versi, Dr. Badi, Dr. Sun, Dr. Abdel‐Aziz and Mrs. Bonatti have nothing to declare.
Figures



References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
