Iron Chelation Therapy Elicits Innate Immune Control of Metastatic Ovarian Cancer
- PMID: 39073085
- PMCID: PMC11452292
- DOI: 10.1158/2159-8290.CD-23-1451
Iron Chelation Therapy Elicits Innate Immune Control of Metastatic Ovarian Cancer
Abstract
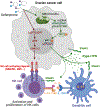
Iron accumulation in tumors contributes to disease progression and chemoresistance. Although targeting this process can influence various hallmarks of cancer, the immunomodulatory effects of iron chelation in the tumor microenvironment are unknown. Here, we report that treatment with deferiprone, an FDA-approved iron chelator, unleashes innate immune responses that restrain ovarian cancer. Deferiprone reprogrammed ovarian cancer cells toward an immunostimulatory state characterized by the production of type-I IFN and overexpression of molecules that activate NK cells. Mechanistically, these effects were driven by innate sensing of mitochondrial DNA in the cytosol and concomitant activation of nuclear DNA damage responses triggered upon iron chelation. Deferiprone synergized with chemotherapy and prolonged the survival of mice with ovarian cancer by bolstering type-I IFN responses that drove NK cell-dependent control of metastatic disease. Hence, iron chelation may represent an alternative immunotherapeutic strategy for malignancies that are refractory to current T-cell-centric modalities. Significance: This study uncovers that targeting dysregulated iron accumulation in ovarian tumors represents a major therapeutic opportunity. Iron chelation therapy using an FDA-approved agent causes immunogenic stress responses in ovarian cancer cells that delay metastatic disease progression and enhance the effects of first-line chemotherapy. See related commentary by Bell and Zou, p. 1771.
©2024 American Association for Cancer Research.
Conflict of interest statement
Conflicts of Interests Statement:
J.R.C.-R. holds patents on the use immune modulators for ovarian cancer treatment and serves as scientific consultant for Moderna, Immagene B.V., Autoimmunity Biologic Solutions, Inc., and Emerald Bioventures, LLC. J.R.C.-R also holds stock options in Vescor Therapeutics. D.Z. reports institutional grants from Merck, Genentech, AstraZeneca, Plexxikon, and Synthekine, and personal fees from AstraZeneca, Xencor, Memgen, Takeda, Synthekine, Immunos, Tessa Therapeutics, Miltenyi, and Calidi Biotherapeutics. D.Z. owns a patent on use of oncolytic Newcastle Disease Virus for cancer therapy. L.G. is/has been holding research contracts with Lytix Biopharma, Promontory and Onxeo, has received consulting/advisory honoraria from Boehringer Ingelheim, AstraZeneca, OmniSEQ, Onxeo, The Longevity Labs, Inzen, Imvax, Sotio, Promontory, Noxopharm, EduCom, and the Luke Heller TECPR2 Foundation, and holds Promontory stock options. All other authors declare no potential conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
- NCC-19112605/National Cancer Center, Korea
- R01 CA249054/CA/NCI NIH HHS/United States
- R01 CA282072/CA/NCI NIH HHS/United States
- Postdoctoral Fellowship Award/Cancer Research Institute (CRI)
- 101067835/Marie Skłodowska-Curie grant
- F31 CA257631/CA/NCI NIH HHS/United States
- N/A/The Sigrid Jusélius Foundation
- MSIT 2020R1C1C1010303/National Research Foundation of Korea (NRF)
- 22-40-15-HWAN/American Association for Cancer Research (AACR)
- T32 5T32AI134632-02/National Institutes of Health (NIH)
- W81XWH2010191/U.S. Department of Defense (DOD)
- R01 CA271915/CA/NCI NIH HHS/United States
- T32 AI134632/AI/NIAID NIH HHS/United States
- SU2C-AACR-IRG-03-16/Stand Up To Cancer (SU2C)
- R01 NS114653/NS/NINDS NIH HHS/United States
- 344697l/PerMed JTC2020 PARIS/Academy of Finland
- K. Albin Johansson Cancer Research Fellowship/Foundation for the Finnish Cancer Institute
- R01 CA271619/CA/NCI NIH HHS/United States
- BC180476P1/U.S. Department of Defense (DOD)
- N/A/The Ovarian Cancer Research Alliance
- N/A/The Cancer Foundation Finland
LinkOut - more resources
Full Text Sources
Medical

