The RPD3L deacetylation complex is required for facultative heterochromatin repression in Neurospora crassa
- PMID: 39074265
- PMCID: PMC11317574
- DOI: 10.1073/pnas.2404770121
The RPD3L deacetylation complex is required for facultative heterochromatin repression in Neurospora crassa
Abstract
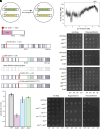
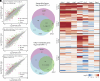
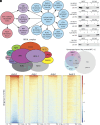
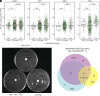
Repression of facultative heterochromatin is essential for developmental processes in numerous organisms. Methylation of histone H3 lysine 27 (H3K27) by Polycomb repressive complex 2 is a prominent feature of facultative heterochromatin in both fungi and higher eukaryotes. Although this methylation is frequently associated with silencing, the detailed mechanism of repression remains incompletely understood. We utilized a forward genetics approach to identify genes required to maintain silencing at facultative heterochromatin genes in Neurospora crassa and identified three previously uncharacterized genes that are important for silencing: sds3 (NCU01599), rlp1 (RPD3L protein 1; NCU09007), and rlp2 (RPD3L protein 2; NCU02898). We found that SDS3, RLP1, and RLP2 associate with N. crassa homologs of the Saccharomyces cerevisiae Rpd3L complex and are required for repression of a subset of H3K27-methylated genes. Deletion of these genes does not lead to loss of H3K27 methylation but increases acetylation of histone H3 lysine 14 at up-regulated genes, suggesting that RPD3L-driven deacetylation is a factor required for silencing of facultative heterochromatin in N. crassa, and perhaps in other organisms.
Keywords: H3K27 methylation; epigenetics; histone modification; polycomb repressive complex; transcriptional silencing.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

