CHIP-mediated ubiquitin degradation of BCAT1 regulates glioma cell proliferation and temozolomide sensitivity
- PMID: 39075053
- PMCID: PMC11286746
- DOI: 10.1038/s41419-024-06938-6
CHIP-mediated ubiquitin degradation of BCAT1 regulates glioma cell proliferation and temozolomide sensitivity
Abstract
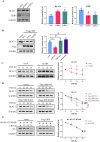
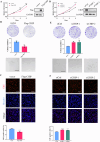
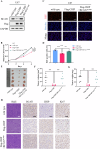
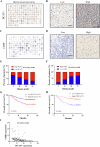
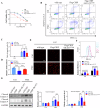
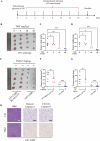
Glioma, a malignant and infiltrative neoplasm of the central nervous system, poses a significant threat due to its high mortality rates. Branched-chain amino acid transaminase 1 (BCAT1), a key enzyme in branched-chain amino acid (BCAA) catabolism, exhibits elevated expression in gliomas and correlates strongly with poor prognosis. Nonetheless, the regulatory mechanisms underlying this increased BCAT1 expression remains incompletely understood. In this study, we reveal that ubiquitination at Lys360 facilitates BCAT1 degradation, with low ubiquitination levels contributing to high BCAT1 expression in glioma cells. The Carboxyl terminus of Hsc70-interacting protein (CHIP), an E3 ubiquitin ligase, interacts with BCAT1 via its coiled-coil (CC) domain, promoting its K48-linkage ubiquitin degradation through proteasomal pathway. Moreover, CHIP-mediated BCAT1 degradation induces metabolic reprogramming, and impedes glioma cell proliferation and tumor growth both in vitro and in vivo. Furthermore, a positive correlation is observed between low CHIP expression, elevated BCAT1 levels, and unfavorable prognosis among glioma patients. Additionally, we show that the CHIP/BCAT1 axis enhances glioma sensitivity to temozolomide by reducing glutathione (GSH) synthesis and increasing oxidative stress. These findings underscore the critical role of CHIP/BCAT1 axis in glioma cell proliferation and temozolomide sensitivity, highlighting its potential as a diagnostic marker and therapeutic target in glioma treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

