Widespread exposure to SARS-CoV-2 in wildlife communities
- PMID: 39075057
- PMCID: PMC11286844
- DOI: 10.1038/s41467-024-49891-w
Widespread exposure to SARS-CoV-2 in wildlife communities
Erratum in
-
Author Correction: Widespread exposure to SARS-CoV-2 in wildlife communities.Nat Commun. 2024 Aug 13;15(1):6940. doi: 10.1038/s41467-024-51220-0. Nat Commun. 2024. PMID: 39138180 Free PMC article. No abstract available.
Abstract
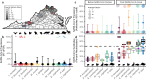
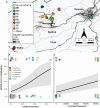
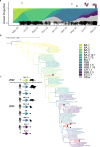
Pervasive SARS-CoV-2 infections in humans have led to multiple transmission events to animals. While SARS-CoV-2 has a potential broad wildlife host range, most documented infections have been in captive animals and a single wildlife species, the white-tailed deer. The full extent of SARS-CoV-2 exposure among wildlife communities and the factors that influence wildlife transmission risk remain unknown. We sampled 23 species of wildlife for SARS-CoV-2 and examined the effects of urbanization and human use on seropositivity. Here, we document positive detections of SARS-CoV-2 RNA in six species, including the deer mouse, Virginia opossum, raccoon, groundhog, Eastern cottontail, and Eastern red bat between May 2022-September 2023 across Virginia and Washington, D.C., USA. In addition, we found that sites with high human activity had three times higher seroprevalence than low human-use areas. We obtained SARS-CoV-2 genomic sequences from nine individuals of six species which were assigned to seven Pango lineages of the Omicron variant. The close match to variants circulating in humans at the time suggests at least seven recent human-to-animal transmission events. Our data support that exposure to SARS-CoV-2 has been widespread in wildlife communities and suggests that areas with high human activity may serve as points of contact for cross-species transmission.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
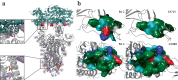
References
-
- World Health Organization. WHO COVID-19 Dashboard.https://covid19.who.int/ (2020).
MeSH terms
Substances
Supplementary concepts
Grants and funding
- AP23OA000000C004/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
- AP23OA000000C004/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
- AP23OA000000C004/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
- AP23OA000000C004/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
- DEB-1911853/National Science Foundation (NSF)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

