Hilbert-curve assisted structure embedding method
- PMID: 39075547
- PMCID: PMC11285582
- DOI: 10.1186/s13321-024-00850-z
Hilbert-curve assisted structure embedding method
Abstract
Motivation: Chemical space embedding methods are widely utilized in various research settings for dimensional reduction, clustering and effective visualization. The maps generated by the embedding process can provide valuable insight to medicinal chemists in terms of the relationships between structural, physicochemical and biological properties of compounds. However, these maps are known to be difficult to interpret, and the ''landscape'' on the map is prone to ''rearrangement'' when embedding different sets of compounds.
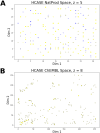
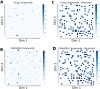
Results: In this study we present the Hilbert-Curve Assisted Space Embedding (HCASE) method which was designed to create maps by organizing structures according to a logic familiar to medicinal chemists. First, a chemical space is created with the help of a set of ''reference scaffolds''. These scaffolds are sorted according to the medicinal chemistry inspired Scaffold-Key algorithm found in prior art. Next, the ordered scaffolds are mapped to a line which is folded into a higher dimensional (here: 2D) space. The intricately folded line is referred to as a pseudo-Hilbert-Curve. The embedding of a compound happens by locating its most similar reference scaffold in the pseudo-Hilbert-Curve and assuming the respective position. Through a series of experiments, we demonstrate the properties of the maps generated by the HCASE method. Subjects of embeddings were compounds of the DrugBank and CANVASS libraries, and the chemical spaces were defined by scaffolds extracted from the ChEMBL database.
Scientific contribution: The novelty of HCASE method lies in generating robust and intuitive chemical space embeddings that are reflective of a medicinal chemist's reasoning, and the precedential use of space filling (Hilbert) curve in the process.
Availability: https://github.com/ncats/hcase.
Keywords: Chemical space embedding; Clustering; Dimension reduction; HCASE; Hilbert-curve; Scaffold-Keys.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Hotelling H (1933) Analysis of a complex of statistical variables into principal components. J Educ. 10.1037/h007132510.1037/h0071325 - DOI
-
- Quist M, Yona G (2004) Distributional scaling: an algorithm for structure-preserving embedding of metric and nonmetric spaces. J Mach Learn Res 5:399–420
-
- L. van der Maaten, “Learning a Parametric Embedding by Preserving Local Structure,” in Proceedings of the Twelth International Conference on Artificial Intelligence and Statistics, D. van Dyk and M. Welling, Eds., in Proceedings of Machine Learning Research, vol. 5. Hilton Clearwater Beach Resort, Clearwater Beach, Florida USA: PMLR, 2009, pp. 384–391.
-
- J. M. Leland McInnes, John Healy. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction.
-
- Kohonen T (1991) Self-organizing maps ophmization approaches. In: Kohonen T, Mäkisara K, Simula O, Kangas J (eds) Artificial Neural Networks. North-Holland, Amsterdam
LinkOut - more resources
Full Text Sources

