TIPRL Regulates Stemness and Survival in Lung Cancer Stem Cells through CaMKK2-CaMK4-CREB Feedback Loop Activation
- PMID: 39076120
- PMCID: PMC11423089
- DOI: 10.1002/advs.202406309
TIPRL Regulates Stemness and Survival in Lung Cancer Stem Cells through CaMKK2-CaMK4-CREB Feedback Loop Activation
Abstract
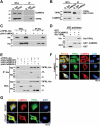
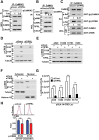
Frequent recurrence and metastasis caused by cancer stem cells (CSCs) are major challenges in lung cancer treatment. Therefore, identifying and characterizing specific CSC targets are crucial for the success of prospective targeted therapies. In this study, it is found that upregulated TOR Signaling Pathway Regulator-Like (TIPRL) in lung CSCs causes sustained activation of the calcium/calmodulin-dependent protein kinase kinase 2 (CaMKK2) signaling pathway by binding to CaMKK2, thereby maintaining stemness and survival. CaMKK2-mediated activation of CaM kinase 4 (CaMK4) leads to phosphorylation of cAMP response element-binding protein (CREB) at Ser129 and Ser133, which is necessary for its maximum activation and the downstream constitutive expression of its target genes (Bcl2 and HMG20A). TIPRL depletion sensitizes lung CSCs to afatinib-induced cell death and reduces distal metastasis of lung cancer in vivo. It is determined that CREB activates the transcription of TIPRL in lung CSCs. The positive feedback loop consisting of CREB and TIPRL induces the sustained activation of the CaMKK2-CaMK4-CREB axis as a driving force and upregulates the expression of stemness- and survival-related genes, promoting tumorigenesis in patients with lung cancer. Thus, TIPRL and the CaMKK2 signaling axis may be promising targets for overcoming drug resistance and reducing metastasis in lung cancer.
Keywords: CaMKK2‐CaMK4‐CREB feedback loop activation; TIPRL; cancer stem cell; drug resistance; lung cancer; metastasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A., CA Cancer J. Clin. 2021, 71, 7. - PubMed
-
- Herbst R. S., Morgensztern D., Boshoff C., Nature 2018, 553, 446. - PubMed
-
- Lee Y. T., Tan Y. J., Oon C. E., Eur. J. Pharmacol. 2018, 834, 188. - PubMed
-
- Krause D. S., Van Etten R. A., N. Engl. J. Med. 2005, 353, 172. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
