Generation of multimillion chemical space based on the parallel Groebke-Blackburn-Bienaymé reaction
- PMID: 39076290
- PMCID: PMC11285076
- DOI: 10.3762/bjoc.20.143
Generation of multimillion chemical space based on the parallel Groebke-Blackburn-Bienaymé reaction
Abstract
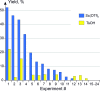
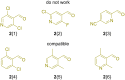
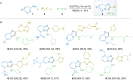
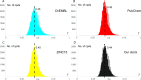
Parallel Groebke-Blackburn-Bienaymé reaction was evaluated as a source of multimillion chemically accessible chemical space. Two most popular classical protocols involving the use of Sc(OTf)3 and TsOH as the catalysts were tested on a broad substrate scope, and prevalence of the first method was clearly demonstrated. Furthermore, the scope and limitations of the procedure were established. A model 790-member library was obtained with 85% synthesis success rate. These results were used to generate a 271-Mln. readily accessible (REAL) heterocyclic chemical space mostly containing unique chemotypes, which was confirmed by comparative analysis with commercially available compound collections. Meanwhile, this chemical space contained 432 compounds that already showed biological activity according to the ChEMBL database.
Keywords: fused rings; heterocycles; imidazoles; isonitrile; multicomponent reactions.
Copyright © 2024, Govor et al.
Figures




References
-
- John S E, Gulati S, Shankaraiah N. Org Chem Front. 2021;8:4237–4287. doi: 10.1039/d0qo01480j. - DOI
LinkOut - more resources
Full Text Sources
