Quantification of Epicardial Adipose Tissue Volume and Attenuation for Cardiac CT Scans Using Deep Learning in a Single Multi-Task Framework
- PMID: 39076659
- PMCID: PMC11270472
- DOI: 10.31083/j.rcm2312412
Quantification of Epicardial Adipose Tissue Volume and Attenuation for Cardiac CT Scans Using Deep Learning in a Single Multi-Task Framework
Abstract
Background: Recent studies have shown that epicardial adipose tissue (EAT) is an independent atrial fibrillation (AF) prognostic marker and has influence on the myocardial function. In computed tomography (CT), EAT volume (EATv) and density (EATd) are parameters that are often used to quantify EAT. While increased EATv has been found to correlate with the prevalence and the recurrence of AF after ablation therapy, higher EATd correlates with inflammation due to arrest of lipid maturation and with high risk of plaque presence and plaque progression. Automation of the quantification task diminishes the variability in readings introduced by different observers in manual quantification and results in high reproducibility of studies and less time-consuming analysis. Our objective is to develop a fully automated quantification of EATv and EATd using a deep learning (DL) framework.
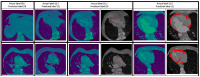
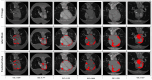
Methods: We proposed a framework that consists of image classification and segmentation DL models and performs the task of selecting images with EAT from all the CT images acquired for a patient, and the task of segmenting the EAT from the output images of the preceding task. EATv and EATd are estimated using the segmentation masks to define the region of interest. For our experiments, a 300-patient dataset was divided into two subsets, each consisting of 150 patients: Dataset 1 (41,979 CT slices) for training the DL models, and Dataset 2 (36,428 CT slices) for evaluating the quantification of EATv and EATd.
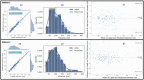
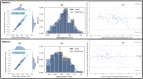
Results: The classification model achieved accuracies of 98% for precision, recall and scores, and the segmentation model achieved accuracies in terms of mean ( std.) and median dice similarity coefficient scores of 0.844 ( 0.19) and 0.84, respectively. Using the evaluation set (Dataset 2), our approach resulted in a Pearson correlation coefficient of 0.971 ( = 0.943) between the label and predicted EATv, and the correlation coefficient of 0.972 ( = 0.945) between the label and predicted EATd.
Conclusions: We proposed a framework that provides a fast and robust strategy for accurate EAT segmentation, and volume (EATv) and attenuation (EATd) quantification tasks. The framework will be useful to clinicians and other practitioners for carrying out reproducible EAT quantification at patient level or for large cohorts and high-throughput projects.
Keywords: CT; EAT; attenuation; deep learning; density; epicardial adipose tissue; volume.
Copyright: © 2022 The Author(s). Published by IMR Press.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- van Rosendael AR, Dimitriu-Leen AC, van Rosendael PJ, Leung M, Smit JM, Saraste A, et al. Association between Posterior Left Atrial Adipose Tissue Mass and Atrial Fibrillation. Circulation: Arrhythmia and Electrophysiology . 2017;10:e004614. - PubMed
-
- Mazurek T, Zhang L, Zalewski A, Mannion JD, Diehl JT, Arafat H, et al. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation . 2003;108:2460–2466. - PubMed
-
- Benjamin EJ, Levy D, Vaziri SM, D’Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. The Journal of the American Medical Association . 1994;271:840–844. - PubMed
-
- Rosengren A, Hauptman PJ, Lappas G, Olsson L, Wilhelmsen L, Swedberg K. Big men and atrial fibrillation: effects of body size and weight gain on risk of atrial fibrillation in men. European Heart Journal . 2009;30:1113–1120. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

