Genomic Discovery and Structure-Activity Exploration of a Novel Family of Enzyme-Activated Covalent Cyclin-Dependent Kinase Inhibitors
- PMID: 39078366
- PMCID: PMC11320645
- DOI: 10.1021/acs.jmedchem.4c01095
Genomic Discovery and Structure-Activity Exploration of a Novel Family of Enzyme-Activated Covalent Cyclin-Dependent Kinase Inhibitors
Abstract
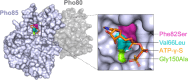
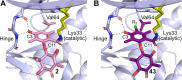
Fungi have historically been the source of numerous important medicinal compounds, but full exploitation of their genetic potential for drug development has been hampered in traditional discovery paradigms. Here we describe a radically different approach, top-down drug discovery (TD3), starting with a massive digital search through a database of over 100,000 fully genomicized fungi to identify loci encoding molecules with a predetermined human target. We exemplify TD3 by the selection of cyclin-dependent kinases (CDKs) as targets and the discovery of two molecules, 1 and 2, which inhibit therapeutically important human CDKs. 1 and 2 exhibit a remarkable mechanism, forming a site-selective covalent bond to the CDK active site Lys. We explored the structure-activity relationship via semi- and total synthesis, generating an analog, 43, with improved kinase selectivity, bioavailability, and efficacy. This work highlights the power of TD3 to identify mechanistically and structurally novel molecules for the development of new medicines.
Conflict of interest statement
The authors declare the following competing financial interest(s): All authors are current or former employees of LifeMine Therapeutics.
Figures













References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

