The circadian rhythm: A key variable in aging?
- PMID: 39078410
- PMCID: PMC11561671
- DOI: 10.1111/acel.14268
The circadian rhythm: A key variable in aging?
Abstract
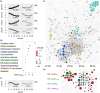
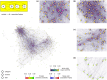
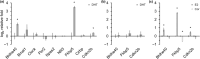
The determination of age-related transcriptional changes may contribute to the understanding of health and life expectancy. The broad application of results from age cohorts may have limitations. Altering sample sizes per time point or sex, using a single mouse strain or tissue, a limited number of replicates, or omitting the middle of life can bias the surveys. To achieve higher general validity and to identify less distinctive players, bulk RNA sequencing of a mouse cohort, including seven organs of two strains from both sexes of 5 ages, was performed. Machine learning by bootstrapped variable importance and selection methodology (Boruta) was used to identify common aging features where the circadian rhythms (CiR) transcripts appear as promising age markers in an unsupervised analysis. Pathways of 11 numerically analyzed local network clusters were affected and classified into four major gene expression profiles, whereby CiR and proteostasis candidates were particularly conspicuous with partially opposing changes. In a data-based interaction association network, the CiR-proteostasis axis occupies an exposed central position, highlighting its relevance. The computation of 11,830 individual transcript associations provides potential superordinate contributors, such as hormones, to age-related changes, as in CiR. In hormone-sensitive LNCaP cells, short-term supraphysiologic levels of the sex hormones dihydrotestosterone or estradiol increase the expression of the CiR transcript Bhlhe40 and the associated senescence regulator Cdkn2b (p15). According to these findings, the bilateral dysregulation of CiR appears as a fundamental protagonist of aging, whose transcripts could serve as a biological marker and its restoration as a therapeutic opportunity.
Keywords: Bhlhe40 (Dec1); aging hallmarks; circadian rhythms; interaction network; machine learning; mouse cohort; organ strain sex; sex hormones.
© 2024 The Author(s). Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Artero‐Castro, A. , Callejas, F. B. , Castellvi, J. , Kondoh, H. , Carnero, A. , Fernandez‐Marcos, P. J. , Serrano, M. , Ramón y Cajal, S. , & Lleonart, M. E. (2009). Cold‐inducible RNA‐binding protein bypasses replicative senescence in primary cells through extracellular signal‐regulated kinase 1 and 2 activation. Molecular and Cellular Biology, 29(7), 1855–1868. 10.1128/MCB.01386-08 - DOI - PMC - PubMed
-
- Cole, J. J. , Robertson, N. A. , Rather, M. I. , Thomson, J. P. , McBryan, T. , Sproul, D. , Wang, T. , Brock, C. , Clark, W. , Ideker, T. , Meehan, R. R. , Miller, R. A. , Brown‐Borg, H. M. , & Adams, P. D. (2017). Diverse interventions that extend mouse lifespan suppress shared age‐associated epigenetic changes at critical gene regulatory regions. Genome Biology, 18(1), 58. 10.1186/s13059-017-1185-3 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

