Congenital Hyperinsulinism and Novel KDM6A Duplications -Resolving Pathogenicity With Genome and Epigenetic Analyses
- PMID: 39078990
- PMCID: PMC12012811
- DOI: 10.1210/clinem/dgae524
Congenital Hyperinsulinism and Novel KDM6A Duplications -Resolving Pathogenicity With Genome and Epigenetic Analyses
Abstract
Context: Hyperinsulinemic hypoglycemia (HI) can be the presenting feature of Kabuki syndrome (KS), which is caused by loss-of-function variants in KMT2D or KDM6A. As these genes play a critical role in maintaining methylation status in chromatin, individuals with pathogenic variants have a disease-specific epigenomic profile-an episignature.
Objective: We evaluated the pathogenicity of 3 novel partial KDM6A duplications identified in 3 individuals presenting with neonatal-onset HI without typical features of KS at the time of genetic testing.
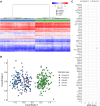
Methods: Three different partial KDM6A duplications were identified by routine targeted next-generation sequencing for HI and initially classified as variants of uncertain significance (VUS) as their location, and hence their impact on the gene, was not known. Whole-genome sequencing (WGS) was undertaken to map the breakpoints of the duplications with DNA methylation profiling performed in 2 individuals to investigate the presence of a KS-specific episignature.
Results: WGS confirmed the duplication in proband 1 as pathogenic as it caused a frameshift in the normal copy of the gene leading to a premature termination codon. The duplications identified in probands 2 and 3 did not alter the reading frame, and therefore their significance remained uncertain after WGS. Subsequent DNA methylation profiling identified a KS-specific episignature in proband 2 but not in proband 3.
Conclusion: Our findings confirm a role for KDM6A partial gene duplications in the etiology of KS and highlight the importance of performing in-depth molecular genetic analysis to properly assess the clinical significance of VUS' in the KDM6A gene.
Keywords: KDM6A; DNA methylation; Kabuki syndrome; congenital hyperinsulinism; episignature; whole-genome sequencing.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Endocrine Society.
Figures

References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

