Rapid iPSC inclusionopathy models shed light on formation, consequence, and molecular subtype of α-synuclein inclusions
- PMID: 39079530
- PMCID: PMC11377155
- DOI: 10.1016/j.neuron.2024.06.002
Rapid iPSC inclusionopathy models shed light on formation, consequence, and molecular subtype of α-synuclein inclusions
Erratum in
-
Rapid iPSC inclusionopathy models shed light on formation, consequence, and molecular subtype of α-synuclein inclusions.Neuron. 2025 Feb 19;113(4):637. doi: 10.1016/j.neuron.2025.01.018. Epub 2025 Feb 1. Neuron. 2025. PMID: 39894019 Free PMC article. No abstract available.
Abstract
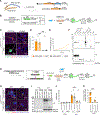
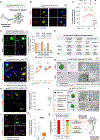
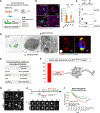
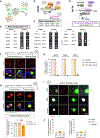
The heterogeneity of protein-rich inclusions and its significance in neurodegeneration is poorly understood. Standard patient-derived iPSC models develop inclusions neither reproducibly nor in a reasonable time frame. Here, we developed screenable iPSC "inclusionopathy" models utilizing piggyBac or targeted transgenes to rapidly induce CNS cells that express aggregation-prone proteins at brain-like levels. Inclusions and their effects on cell survival were trackable at single-inclusion resolution. Exemplar cortical neuron α-synuclein inclusionopathy models were engineered through transgenic expression of α-synuclein mutant forms or exogenous seeding with fibrils. We identified multiple inclusion classes, including neuroprotective p62-positive inclusions versus dynamic and neurotoxic lipid-rich inclusions, both identified in patient brains. Fusion events between these inclusion subtypes altered neuronal survival. Proteome-scale α-synuclein genetic- and physical-interaction screens pinpointed candidate RNA-processing and actin-cytoskeleton-modulator proteins like RhoA whose sequestration into inclusions could enhance toxicity. These tractable CNS models should prove useful in functional genomic analysis and drug development for proteinopathies.
Keywords: CRISPR screen; Lewy body; Parkinson’s disease; Rab protein; RhoA; actin cytoskeleton; aggregation; dementia with Lewy bodies; glia; iPSC; inclusion; lipid; neurodegeneration; neuron; p62; piggyBac; proximity labeling; synucleinopathy; ubiquitin; α-synuclein.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests V.K. is a cofounder of and senior advisor to DaCapo Brainscience and Yumanity Therapeutics, companies focused on CNS diseases. C.Y.C. and X.J. contributed to this work as employees of Yumanity Therapeutics. T.I. and Y.K. contributed to this work as employees of Nikon Corporation. I.L., A.N., J. Sandoe, and V.K. are inventors on a patent application filed by Brigham and Women’s Hospital related to the induced inclusion iPSC models.
Figures



References
-
- Van der Perren A, Gelders G, Fenyi A, Bousset L, Brito F, Peelaerts W, Van den Haute C, Gentleman S, Melki R, and Baekelandt V (2020). The structural differences between patient-derived α-synuclein strains dictate characteristics of Parkinson’s disease, multiple system atrophy and dementia with Lewy bodies. Acta Neuropathol 139, 977–1000. 10.1007/s00401-020-02157-3. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

