Proteogenomic network analysis reveals dysregulated mechanisms and potential mediators in Parkinson's disease
- PMID: 39080267
- PMCID: PMC11289099
- DOI: 10.1038/s41467-024-50718-x
Proteogenomic network analysis reveals dysregulated mechanisms and potential mediators in Parkinson's disease
Abstract
Parkinson's disease is highly heterogeneous across disease symptoms, clinical manifestations and progression trajectories, hampering the identification of therapeutic targets. Despite knowledge gleaned from genetics analysis, dysregulated proteome mechanisms stemming from genetic aberrations remain underexplored. In this study, we develop a three-phase system-level proteogenomic analytical framework to characterize disease-associated proteins and dysregulated mechanisms. Proteogenomic analysis identified 577 proteins that enrich for Parkinson's disease-related pathways, such as cytokine receptor interactions and lysosomal function. Converging lines of evidence identified nine proteins, including LGALS3, CSNK2A1, SMPD3, STX4, APOA2, PAFAH1B3, LDLR, HSPB1, BRK1, with potential roles in disease pathogenesis. This study leverages the largest population-scale proteomics dataset, the UK Biobank Pharma Proteomics Project, to characterize genetically-driven protein disturbances associated with Parkinson's disease. Taken together, our work contributes to better understanding of genome-proteome dynamics in Parkinson's disease and sets a paradigm to identify potential indirect mediators connected to GWAS signals for complex neurodegenerative disorders.
© 2024. The Author(s).
Conflict of interest statement
A.D.T., D.T.T., L.H., B.S., C.D.W., and S.L. are full-time employees of Janssen R&D and are stockholders of Johnson & Johnson.
Figures

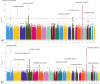

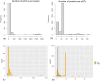
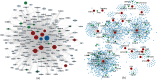


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

