Multi-generational benefits of genetic rescue
- PMID: 39080286
- PMCID: PMC11289468
- DOI: 10.1038/s41598-024-67033-6
Multi-generational benefits of genetic rescue
Abstract
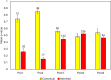
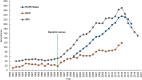
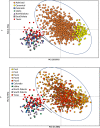
Genetic rescue-an increase in population fitness following the introduction of new alleles-has been proven to ameliorate inbreeding depression in small, isolated populations, yet is rarely applied as a conservation tool. A lingering question regarding genetic rescue in wildlife conservation is how long beneficial effects persist in admixed populations. Using data collected over 40 years from 1192 endangered Florida panthers (Puma concolor coryi) across nine generations, we show that the experimental genetic rescue implemented in 1995-via the release of eight female pumas from Texas-alleviated morphological, genetic, and demographic correlates of inbreeding depression, subsequently preventing extirpation of the population. We present unequivocal evidence, for the first time in any terrestrial vertebrate, that genetic and phenotypic benefits of genetic rescue remain in this population after five generations of admixture, which helped increase panther abundance (> fivefold) and genetic effective population size (> 20-fold). Additionally, even with extensive admixture, microsatellite allele frequencies in the population continue to support the distinctness of Florida panthers from other North American puma populations, including Texas. Although threats including habitat loss, human-wildlife conflict, and infectious diseases are challenges to many imperiled populations, our results suggest genetic rescue can serve as an effective, multi-generational tool for conservation of small, isolated populations facing extinction from inbreeding.
Keywords: Puma concolor; Endangered species conservation; Fitness; Florida panther; Genetic rescue; Inbreeding depression.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Genetic restoration of the Florida panther.Science. 2010 Sep 24;329(5999):1641-5. doi: 10.1126/science.1192891. Science. 2010. PMID: 20929847 Free PMC article.
-
A cat's tale: the impact of genetic restoration on Florida panther population dynamics and persistence.J Anim Ecol. 2013 May;82(3):608-20. doi: 10.1111/1365-2656.12033. Epub 2012 Dec 17. J Anim Ecol. 2013. PMID: 23252671
-
Intentional genetic introgression influences survival of adults and subadults in a small, inbred felid population.J Anim Ecol. 2011 Sep;80(5):958-67. doi: 10.1111/j.1365-2656.2011.01809.x. Epub 2011 Feb 21. J Anim Ecol. 2011. PMID: 21338353 Free PMC article.
-
Genetic rescue to the rescue.Trends Ecol Evol. 2015 Jan;30(1):42-9. doi: 10.1016/j.tree.2014.10.009. Epub 2014 Nov 27. Trends Ecol Evol. 2015. PMID: 25435267 Review.
-
Conservation and Genetics.Yale J Biol Med. 2018 Dec 21;91(4):491-501. eCollection 2018 Dec. Yale J Biol Med. 2018. PMID: 30588214 Free PMC article. Review.
Cited by
-
A chromosome-level genome assembly of the mountain lion, Puma concolor.J Hered. 2025 Jul 21;116(4):479-487. doi: 10.1093/jhered/esae063. J Hered. 2025. PMID: 39506490 Free PMC article.
-
Genetic rescue of Florida panthers reduced homozygosity but did not swamp ancestral genotypes.Proc Natl Acad Sci U S A. 2025 Aug 5;122(31):e2410945122. doi: 10.1073/pnas.2410945122. Epub 2025 Jul 28. Proc Natl Acad Sci U S A. 2025. PMID: 40720660 Free PMC article.
-
Sexual selection matters in genetic rescue, but productivity benefits fade over time: a multi-generation experiment to inform conservation.Proc Biol Sci. 2025 Jan;292(2039):20242374. doi: 10.1098/rspb.2024.2374. Epub 2025 Jan 29. Proc Biol Sci. 2025. PMID: 39876725 Free PMC article.
-
Whole Genomes Inform Genetic Rescue Strategy for Montane Red Foxes in North America.Mol Biol Evol. 2024 Sep 4;41(9):msae193. doi: 10.1093/molbev/msae193. Mol Biol Evol. 2024. PMID: 39288165 Free PMC article.
References
-
- Gilpin, M. & Soulé, M. E. Conservation Biology: The Science of Scarcity and Diversity 19–34 (Sinauer Associates, 1986).
MeSH terms
LinkOut - more resources
Full Text Sources

