Comprehensive proteomic analysis of the differential expression of 62 proteins following intracortical microelectrode implantation
- PMID: 39080300
- PMCID: PMC11289480
- DOI: 10.1038/s41598-024-68017-2
Comprehensive proteomic analysis of the differential expression of 62 proteins following intracortical microelectrode implantation
Abstract
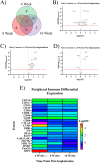
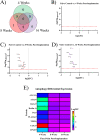
Intracortical microelectrodes (IMEs) are devices designed to be implanted into the cerebral cortex for various neuroscience and neuro-engineering applications. A critical feature of IMEs is their ability to detect neural activity from individual neurons. Currently, IMEs are limited by chronic failure, largely considered to be caused by the prolonged neuroinflammatory response to the implanted devices. Over the past few years, the characterization of the neuroinflammatory response has grown in sophistication, with the most recent advances focusing on mRNA expression following IME implantation. While gene expression studies increase our broad understanding of the relationship between IMEs and cortical tissue, advanced proteomic techniques have not been reported. Proteomic evaluation is necessary to describe the diverse changes in protein expression specific to neuroinflammation, neurodegeneration, or tissue and cellular viability, which could lead to the further development of targeted intervention strategies designed to improve IME functionality. In this study, we have characterized the expression of 62 proteins within 180 μm of the IME implant site at 4-, 8-, and 16-weeks post-implantation. We identified potential targets for immunotherapies, as well as key pathways that contribute to neuronal dieback around the IME implant.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Comprehensive Proteomic Analysis of the Differential Expression of 83 Proteins Following Intracortical Microelectrode Implantation.Res Sq [Preprint]. 2024 Mar 12:rs.3.rs-4039586. doi: 10.21203/rs.3.rs-4039586/v1. Res Sq. 2024. Update in: Sci Rep. 2024 Jul 30;14(1):17596. doi: 10.1038/s41598-024-68017-2. PMID: 38559066 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

