TRPML1 activation ameliorates lysosomal phenotypes in CLN3 deficient retinal pigment epithelial cells
- PMID: 39080379
- PMCID: PMC11289453
- DOI: 10.1038/s41598-024-67479-8
TRPML1 activation ameliorates lysosomal phenotypes in CLN3 deficient retinal pigment epithelial cells
Abstract
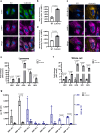
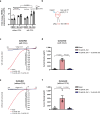
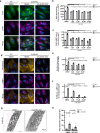
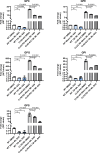
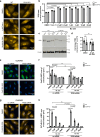
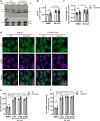
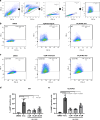
Mutations in the lysosomal membrane protein CLN3 cause Juvenile Neuronal Ceroid Lipofuscinosis (JNCL). Activation of the lysosomal ion channel TRPML1 has previously been shown to be beneficial in several neurodegenerative disease models. Here, we tested whether TRPML1 activation rescues disease-associated phenotypes in CLN3-deficient retinal pigment epithelial (ARPE-19 CLN3-KO) cells. ARPE-19 CLN3-KO cells accumulate LAMP1 positive organelles and show lysosomal storage of mitochondrial ATPase subunit C (SubC), globotriaosylceramide (Gb3), and glycerophosphodiesters (GPDs), whereas lysosomal bis(monoacylglycero)phosphate (BMP/LBPA) lipid levels were significantly decreased. Activation of TRPML1 reduced lysosomal storage of Gb3 and SubC but failed to restore BMP levels in CLN3-KO cells. TRPML1-mediated decrease of storage was TFEB-independent, and we identified TRPML1-mediated enhanced lysosomal exocytosis as a likely mechanism for clearing storage including GPDs. Therefore, ARPE-19 CLN3-KO cells represent a human cell model for CLN3 disease showing many of the described core lysosomal deficits, some of which can be improved using TRPML1 agonists.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

