Comprehensive analysis of circRNA-miRNA-mRNA network related to angiogenesis in recurrent implantation failure
- PMID: 39080700
- PMCID: PMC11290139
- DOI: 10.1186/s12920-024-01944-1
Comprehensive analysis of circRNA-miRNA-mRNA network related to angiogenesis in recurrent implantation failure
Abstract
Background: Abnormal endometrial blood flow causes a decrease in endometrial receptivity and is considered a relatively independent risk factor for recurrent implantation failure (RIF). This study aimed to explore the potentially functional circRNA-miRNA-mRNA network in RIF, and further explore its mechanism.
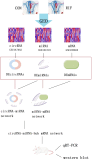
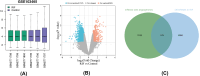
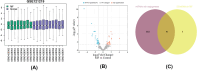
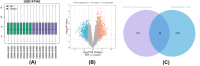
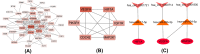
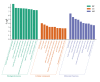
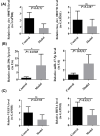
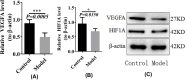
Methods: Datasets were downloaded from the GEO database to identify differentially expressed circRNAs, miRNAs and mRNAs. The circRNA-miRNA-mRNA and PPI networks were constructed using Cytoscape 3.6.0 and the STRING database, the hub genes were identified with the cytoHubba plug-in, and a circRNA-miRNA-hub mRNA regulatory sub-network was constructed. Then, GO and KEGG pathway enrichment analyses of the hub genes were performed to comprehensively analyze the mechanism of hub mRNAs in RIF. Due to the results of circRNAs-miRNAs-hub mRNAs regulatory network, we verified the expression of circRNA_0001721, circRNA_0000714, miR-17-5p, miR-29b-3p, HIF1A and VEGFA in the RIF mouse model by qRT‒PCR and western blotting.
Results: We initially identified 175 DEmRNAs, 48 DEmiRNAs and 56 DEcircRNAs in RIF associated with angiogenesis and constructed a circRNA-miRNA‒mRNA network and PPI network. We further identified six hub genes in the acquired network. Based on these genes, functional enrichment analysis revealed that the HIF-1 signaling pathway plays a vital role in endometrial angiogenesis in RIF. In addition, the interaction networks of circRNA_0001721/miR-17-5p/HIF1A and the circRNA_0000714/miR-29b-3p/VEGFA axis were predicted. In the RIF mouse model, circRNA_0001721, circRNA_0000714, HIF1A and VEGFA were down-regulated, whereas miR-17-5p and miR-29b-3p were up-regulated according to qRT‒PCR and western blotting.
Conclusion: This study revealed that the HIF-1 signaling pathway plays a vital role in endometrial angiogenesis in RIF. The circRNA_0001721/miR-17-5p/HIF1A and circRNA_0000714/miR-29b-3p/VEGFA axes might play a role in the pathogenesis of endometrial angiogenesis in RIF.
Keywords: Angiogenesis; Circular RNA; Endometrial receptivity; Molecular pathogenesis; Recurrent implantation failure; Regulatory network; Signaling pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Construction of Circular RNA-MicroRNA-Messenger RNA Regulatory Network of Recurrent Implantation Failure to Explore Its Potential Pathogenesis.Front Genet. 2021 Feb 16;11:627459. doi: 10.3389/fgene.2020.627459. eCollection 2020. Front Genet. 2021. PMID: 33664765 Free PMC article.
-
Reveal the potential molecular mechanism of circRNA regulating immune-related mRNA through sponge miRNA in the occurrence and immune regulation of papillary thyroid cancer.Ann Med. 2023;55(2):2244515. doi: 10.1080/07853890.2023.2244515. Ann Med. 2023. PMID: 37603701 Free PMC article.
-
Hsa_circ_0038383-mediated competitive endogenous RNA network in recurrent implantation failure.Aging (Albany NY). 2021 Feb 20;13(4):6076-6090. doi: 10.18632/aging.202590. Epub 2021 Feb 20. Aging (Albany NY). 2021. PMID: 33611311 Free PMC article.
-
Network pharmacology-based identification of miRNA expression of Astragalus membranaceus in the treatment of diabetic nephropathy.Medicine (Baltimore). 2022 Feb 4;101(5):e28747. doi: 10.1097/MD.0000000000028747. Medicine (Baltimore). 2022. PMID: 35119030 Free PMC article.
-
MicroRNA Signatures in Endometrial Receptivity-Unlocking Their Role in Embryo Implantation and IVF Success: A Systematic Review.Biomedicines. 2025 May 13;13(5):1189. doi: 10.3390/biomedicines13051189. Biomedicines. 2025. PMID: 40427016 Free PMC article. Review.
Cited by
-
Enhancing Skin Rejuvenation: Using of Engineered Exosome Content Treated With Oleuropein and Fe3O4@CQD/Oleuropein.J Cosmet Dermatol. 2025 Jul;24(7):e70351. doi: 10.1111/jocd.70351. J Cosmet Dermatol. 2025. PMID: 40654202 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

