Targeted dephosphorylation of TFEB promotes its nuclear translocation
- PMID: 39081292
- PMCID: PMC11284556
- DOI: 10.1016/j.isci.2024.110432
Targeted dephosphorylation of TFEB promotes its nuclear translocation
Abstract
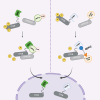
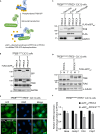
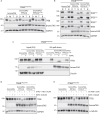
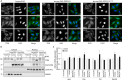
Reversible phosphorylation of the transcription factor EB (TFEB) coordinates cellular responses to metabolic and other stresses. During nutrient replete and stressor-free conditions, phosphorylated TFEB is primarily localized to the cytoplasm. Stressor-mediated reduction of TFEB phosphorylation promotes its nuclear translocation and context-dependent transcriptional activity. In this study, we explored targeted dephosphorylation of TFEB as an approach to activate TFEB in the absence of nutrient deprivation or other cellular stress. Through an induction of proximity between TFEB and several phosphatases using the AdPhosphatase system, we demonstrate targeted dephosphorylation of TFEB in cells. Furthermore, by developing a heterobifunctional molecule BDPIC (bromoTAG-dTAG proximity-inducing chimera), we demonstrate targeted dephosphorylation of TFEB-dTAG through induced proximity to bromoTAG-PPP2CA. Targeted dephosphorylation of TFEB-dTAG by bromoTAG-PPP2CA with BDPIC at the endogenous levels is sufficient to induce nuclear translocation and some transcriptional activity of TFEB.
Keywords: Health sciences; Medical specialty; Medicine; Precision medicine.
© 2024 The Authors.
Conflict of interest statement
University of Dundee is currently in the process of filing a patent application for the bivalent molecule BDPIC that was developed as part of this study. The authors declare no other competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

