MetaGate: Interactive analysis of high-dimensional cytometry data with metadata integration
- PMID: 39081571
- PMCID: PMC11284499
- DOI: 10.1016/j.patter.2024.100989
MetaGate: Interactive analysis of high-dimensional cytometry data with metadata integration
Abstract
Flow cytometry is a powerful technology for high-throughput protein quantification at the single-cell level. Technical advances have substantially increased data complexity, but novel bioinformatical tools often show limitations in statistical testing, data sharing, cross-experiment comparability, or clinical data integration. We developed MetaGate as a platform for interactive statistical analysis and visualization of manually gated high-dimensional cytometry data with integration of metadata. MetaGate provides a data reduction algorithm based on a combinatorial gating system that produces a small, portable, and standardized data file. This is subsequently used to produce figures and statistical analyses through a fast web-based user interface. We demonstrate the utility of MetaGate through a comprehensive mass cytometry analysis of peripheral blood immune cells from 28 patients with diffuse large B cell lymphoma along with 17 healthy controls. Through MetaGate analysis, our study identifies key immune cell population changes associated with disease progression.
Keywords: data analysis; diffuse large B-cell lymphoma; flow cytometry; mass cytometry.
© 2024 The Authors.
Conflict of interest statement
K-J.M. is a consultant at Fate Therapeutics and Vycellix and has research support from Oncopeptides for studies unrelated to this work.
Figures
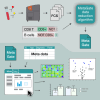


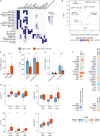

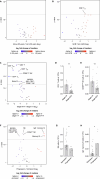
Update of
-
MetaGate: Interactive Analysis of High-Dimensional Cytometry Data with Meta Data Integration.bioRxiv [Preprint]. 2023 Nov 1:2023.10.27.564454. doi: 10.1101/2023.10.27.564454. bioRxiv. 2023. Update in: Patterns (N Y). 2024 May 13;5(7):100989. doi: 10.1016/j.patter.2024.100989. PMID: 37961421 Free PMC article. Updated. Preprint.
References
-
- Cheung M., Campbell J.J., Whitby L., Thomas R.J., Braybrook J., Petzing J. Current trends in flow cytometry automated data analysis software. Cytometry A. 2021;99:1007–1021. - PubMed
-
- Van der Maaten L., Hinton G. Visualizing data using t-SNE. Journal of machine learning research. 2008;9
Grants and funding
LinkOut - more resources
Full Text Sources

