CoPixie, a novel algorithm for single-particle track colocalization, enables efficient quantification of telomerase dynamics at telomeres
- PMID: 39082280
- PMCID: PMC11381360
- DOI: 10.1093/nar/gkae669
CoPixie, a novel algorithm for single-particle track colocalization, enables efficient quantification of telomerase dynamics at telomeres
Abstract
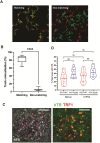
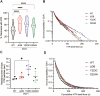
Single-particle imaging and tracking can be combined with colocalization analysis to study the dynamic interactions between macromolecules in living cells. Indeed, single-particle tracking has been extensively used to study protein-DNA interactions and dynamics. Still, unbiased identification and quantification of binding events at specific genomic loci remains challenging. Herein, we describe CoPixie, a new software that identifies colocalization events between a theoretically unlimited number of imaging channels, including single-particle movies. CoPixie is an object-based colocalization algorithm that relies on both pixel and trajectory overlap to determine colocalization between molecules. We employed CoPixie with live-cell single-molecule imaging of telomerase and telomeres, to test the model that cancer-associated POT1 mutations facilitate telomere accessibility. We show that POT1 mutants Y223C, D224N or K90E increase telomere accessibility for telomerase interaction. However, unlike the POT1-D224N mutant, the POT1-Y223C and POT1-K90E mutations also increase the duration of long-lasting telomerase interactions at telomeres. Our data reveal that telomere elongation in cells expressing cancer-associated POT1 mutants arises from the dual impact of these mutations on telomere accessibility and telomerase retention at telomeres. CoPixie can be used to explore a variety of questions involving macromolecular interactions in living cells, including between proteins and nucleic acids, from multicolor single-particle tracks.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Lagache T., Sauvonnet N., Danglot L., Olivo-Marin J.-C.. Statistical analysis of molecule colocalization in bioimaging. Cytometry A. 2015; 87:568–579. - PubMed
-
- Luo F., Qin G., Xia T., Fang X.. Single-molecule imaging of protein interactions and dynamics. Annu. Rev. Anal. Chem. 2020; 13:337–361. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

