A Pan-Cancer Patient-Derived Xenograft Histology Image Repository with Genomic and Pathologic Annotations Enables Deep Learning Analysis
- PMID: 39082680
- PMCID: PMC11217732
- DOI: 10.1158/0008-5472.CAN-23-1349
A Pan-Cancer Patient-Derived Xenograft Histology Image Repository with Genomic and Pathologic Annotations Enables Deep Learning Analysis
Abstract
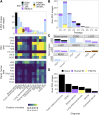
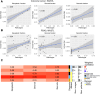
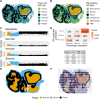
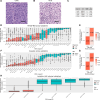
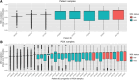
Patient-derived xenografts (PDX) model human intra- and intertumoral heterogeneity in the context of the intact tissue of immunocompromised mice. Histologic imaging via hematoxylin and eosin (H&E) staining is routinely performed on PDX samples, which could be harnessed for computational analysis. Prior studies of large clinical H&E image repositories have shown that deep learning analysis can identify intercellular and morphologic signals correlated with disease phenotype and therapeutic response. In this study, we developed an extensive, pan-cancer repository of >1,000 PDX and paired parental tumor H&E images. These images, curated from the PDX Development and Trial Centers Research Network Consortium, had a range of associated genomic and transcriptomic data, clinical metadata, pathologic assessments of cell composition, and, in several cases, detailed pathologic annotations of neoplastic, stromal, and necrotic regions. The amenability of these images to deep learning was highlighted through three applications: (i) development of a classifier for neoplastic, stromal, and necrotic regions; (ii) development of a predictor of xenograft-transplant lymphoproliferative disorder; and (iii) application of a published predictor of microsatellite instability. Together, this PDX Development and Trial Centers Research Network image repository provides a valuable resource for controlled digital pathology analysis, both for the evaluation of technical issues and for the development of computational image-based methods that make clinical predictions based on PDX treatment studies. Significance: A pan-cancer repository of >1,000 patient-derived xenograft hematoxylin and eosin-stained images will facilitate cancer biology investigations through histopathologic analysis and contributes important model system data that expand existing human histology repositories.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
B.S. White reports grants from the NIH/NCI during the conduct of the study. T. Sheridan reports personal fees from Google outside the submitted work. S.R. Davies reports grants from NIH/NCI U54CA224083 during the conduct of the study. K.W. Evans reports grants from the NCI during the conduct of the study. B.J. Sanderson reports grants from the NIH/NCI during the conduct of the study. M.W. Lloyd reports grants from NCI (HHS—NIH) during the conduct of the study. L.E. Dobrolecki reports grants from the NIH during the conduct of the study, as well as personal fees from StemMed, Ltd. outside the submitted work. B.N. Davis-Dusenbery reports grants and other support from the NCI during the conduct of the study, as well as being an employee and equity holder in Velsera. N. Mitsiades reports grants from the NCI during the conduct of the study. A.L. Welm reports that The University of Utah may license the models described herein to for-profit companies, which may result in tangible property royalties to the university and members of the Welm Labs who developed the models. B.E. Welm reports grants from the NIH/NCI during the conduct of the study; receiving royalties from licenses of previously developed PDX models issued by The University of Utah; and that The University of Utah may issue new licenses in the future at its discretion, which may result in additional royalties to the authors. S. Li reports personal fees from Inotivco outside the submitted work. M.A. Davies reports grants from the NCI during the conduct of the study, as well as personal fees from Roche/Genentech, Pfizer, Novartis, Bristol Myers Squibb, Iovance, and Eisai, grants and personal fees from ABM Therapeutics, and grants from Lead Pharma outside the submitted work. F. Meric-Bernstam reports personal fees from AbbVie, Aduro BioTech Inc., Alkermes, AstraZeneca, Daiichi Sankyo Co., Ltd., Calibr (a division of Scripps Research), Debiopharm, EcoR1 Capital, eFFECTOR Therapeutics, F. Hoffmann-La Roche Ltd., GT Apeiron, Genentech, Inc., Harbinger Health, IBM Watson, Incyte, Infinity Pharmaceuticals, The Jackson Laboratory, KOLON Life Science, LegoChem Bio, Lengo Therapeutics, Menarini Group, OrigiMed, PACT Pharma, Parexel International, Pfizer Inc., Protai Bio Ltd., Samsung Bioepis, Seattle Genetics, Inc., Tallac Therapeutics, Tyra Biosciences, Xencor, Zymeworks, Black Diamond, Biovica, Eisai, FogPharma, Immunomedics, Inflection Biosciences, Karyopharm Therapeutics, Loxo Oncology, Mersana Therapeutics, OnCusp Therapeutics, Puma Biotechnology Inc., Sanofi, Silverback Therapeutics, Spectrum Pharmaceuticals, Theratechnologies, Zentalis and Dava Oncology; grants from Aileron Therapeutics, Inc., AstraZeneca, Bayer Healthcare Pharmaceuticals, Calithera Biosciences Inc., Curis, Inc., CytomX Therapeutics Inc., Daiichi Sankyo Co., Ltd., Debiopharm International, eFFECTOR Therapeutics, Genentech, Inc., Guardant Health, Inc., KLUS Pharma, Takeda Pharmaceuticals, Novartis, Puma Biotechnology, Inc., and Taiho Pharmaceutical Co.; and other support from European Organisation for Research and Treatment of Cancer, European Society for Medical Oncology, Cholangiocarcinoma Foundation, and Dava Oncology outside the submitted work. Y. Xie reports grants from NIH and Cancer Prevention and Research Institute of Texas (CPRIT) during the conduct of the study; grants from NIH and CPRIT outside the submitted work; and being a cofounder of the Adjuvant Genomics, Inc. M.T. Lewis reports grants from the NCI during the conduct of the study, as well as being a founder and limited partner in StemMed Ltd., founder, manager, and general partner in StemMed Holdings LLC, and founder and equity holder in Tvardi Therapeutics Inc. No disclosures were reported by the other authors.
Figures

References
MeSH terms
Grants and funding
- U54 CA233223/CA/NCI NIH HHS/United States
- P50 CA261608/CA/NCI NIH HHS/United States
- U54 CA283766/CA/NCI NIH HHS/United States
- HHSN261201400008C/CA/NCI NIH HHS/United States
- U24 CA224067/CA/NCI NIH HHS/United States
- U54 CA224070/CA/NCI NIH HHS/United States
- HHSN261200800001C/RC/CCR NIH HHS/United States
- HHSN261200800001E/CA/NCI NIH HHS/United States
- U24-CA224067/Cancer Moonshot (Misión contra el Cáncer)
- U54 CA224076/CA/NCI NIH HHS/United States
- U54 CA224065/CA/NCI NIH HHS/United States
- U54 CA233306/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- R01 CA089713/CA/NCI NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- U54 CA224083/CA/NCI NIH HHS/United States
- HHSN261201500003C/CA/NCI NIH HHS/United States
- R01 CA230031/CA/NCI NIH HHS/United States
- HHSN261201500003I/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

